Method for producing novel hipsc by means of sirna introduction
An inducer and pluripotent stem cell technology, applied in the field of new hiPSC production method using miRNA introduction, can solve the problem of hTERTmRNA expression reduction and other problems
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0176]
[0177] (1-1) Production of RGM249shRNA
[0178] According to Stealth RNAi designer (Invitrogen, California, USA), the sequence of RGM249shRNA was designed, and a vector (RGM249shRNA plasmid) for producing RGM249shRNA was produced using BLOCK-it Inducible H1RNAi Entry Vector (Invitrogen, California, USA). This RGM249shRNA is a small RNA designed to induce RNAi against RGM249mRNA in vivo.
[0179] Here, the base sequence of RGM249 mRNA is
[0180] 5’-GGAAAACUAAAAUGAGAGAAUGGGUGUCCAAGAGGACAAGUUCAUGCUCACCCGGUGAUGAGAGUUUGAUUGCAGAAUAAGGCUAGACAAAGGGAAGCUGAACAUGACCAAAGCCAUGUGACAUCGUAUGAUCCUCGAAUCUCACAGUAUCUAUGUAUCUAUAAUCAGAUACAUCCCUAGACUUUCCAGGAAUUCUGGUACUUCACGAGGAUGUGAGAAGACUCUGAACAAAAUAAUACACUGCUCGUG-3’(序列号7)。
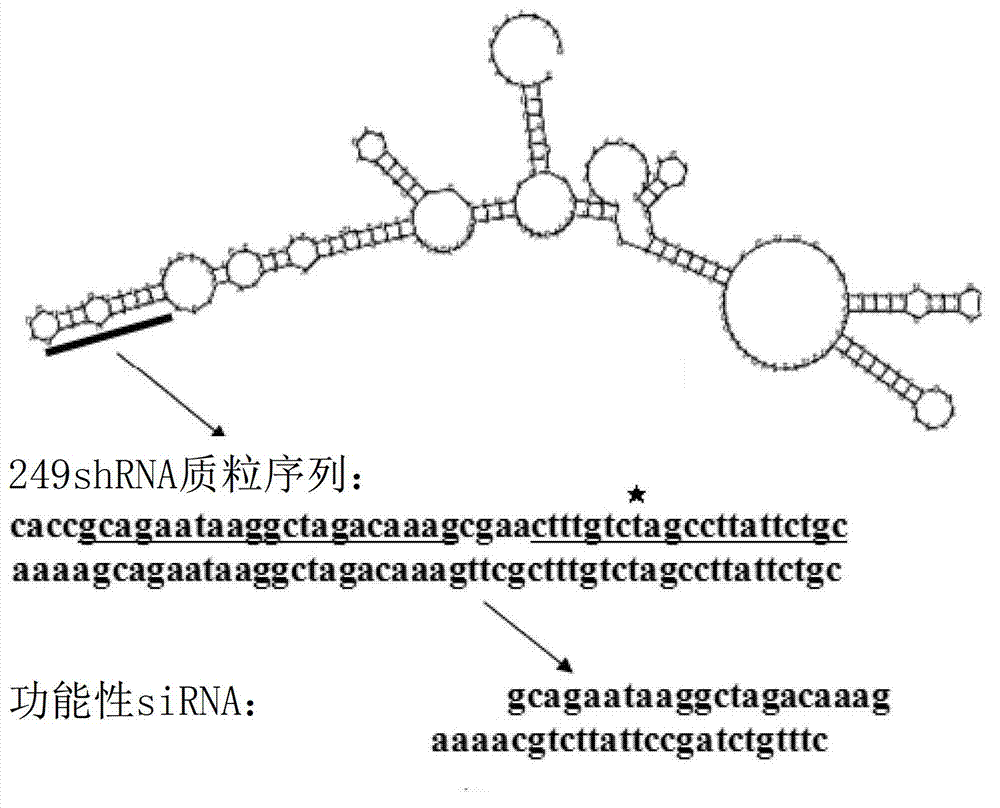
[0181] In the upper strand of the RGM249shRNA plasmid, the sequence encoding RGM249shRNA is
[0182] 5'-CACCGCAGAATAAGGCTAGACAAAGCGAACTTTGTCTAGCCTTATTCTGC-3' (SEQ ID NO: 11). In the lower strand, the sequence forming a double strand with the upper strand is 5'-A...
Embodiment 2
[0202]
[0203] (2-1) Structure of miR-47siRNA, miR-101siRNA, miR-197siRNA
[0204] RGM249 was linked to the pRNAT-U6.1 / neo vector (GenScript USA, NJ, USA). RGM249 mRNA generated by T7 RNA polymerase was digested with RNA endonuclease III (Genlantis, CA, USA). The miRNA was fractionated by mirVANA miRNA Isolation Kit (Ambion, Japan, Tokyo, Japan), and purified by miRNA Isolation Kit (Wako Pure Chemical Industries, Ltd., Tokyo, Japan) using human anti-Ago2 microbeads. In addition, since there was a possibility that the small RNA did not bind to Ago2, purification was not performed using anti-Ago2. The digested small RNAs were cloned using the miRCAT-microRNA Cloning Kit (Integrated DNA Technologies, Iowa, USA), and sequenced using the TOPO vector (Invitrogen, California, USA). Additionally, secondary structures are predicted (http: / / rna.tbi.univie.ac.at / cgi-bin / RNAfold.cgi). Using miRBas, study the sequence identity of small RNAs. From the above results, three miRNAs, miR...
Embodiment 3
[0218]
[0219] (3-1) Subcutaneous administration of siRNA
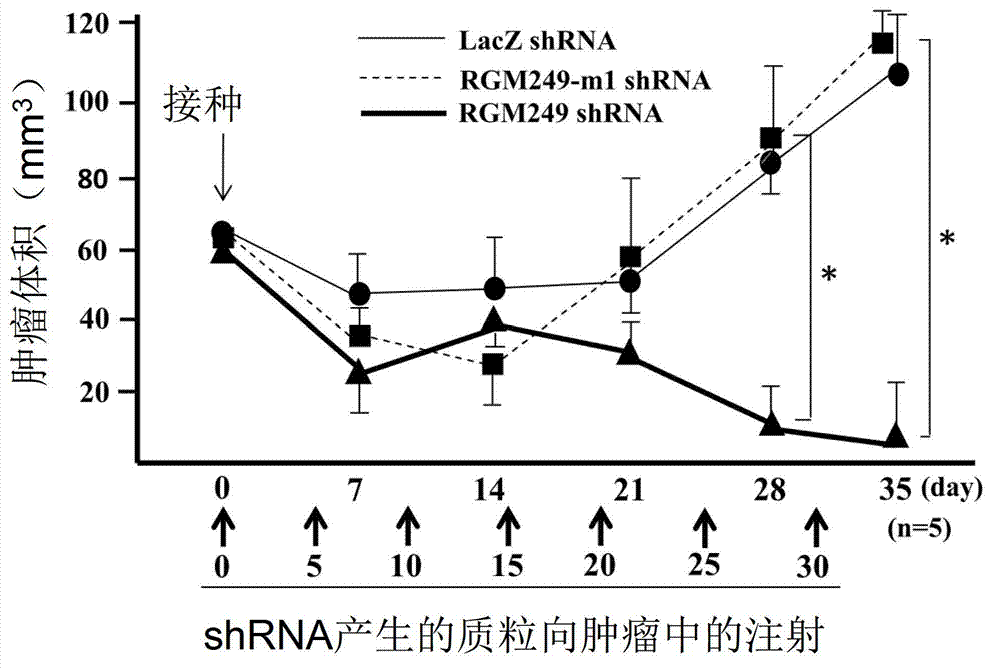
[0220] Subcutaneous administration of 3 siRNA mixtures + DDS showed inhibition of proliferation of HMV-I cells compared to control administration of DDS alone (PTM ). 3 siRNA mixes and DDS were mixed at 100 μM each according to the manufacturer's protocol. The photographs shown in the upper part of FIG. 4 are photographs when a non-RNA-containing collagen (mock) was subcutaneously injected. The photographs shown in the lower part are photographs when 3 siRNA mixtures + DDS were subcutaneously injected. *Indicates that there is a significant difference between the case without RNA and the case with siRNA (P<0.01). Additionally, control mice displayed multiple nodules in the lungs (15.8 ± 1.9 nodules) and intraperitoneal cavity (0.8 ± 0.6 nodules) (Table 2). Several metastatic lesions were observed intraperitoneally as well as extraperitoneally, and subcutaneous infiltration was also observed.
[0221] 【Table 2】 ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com