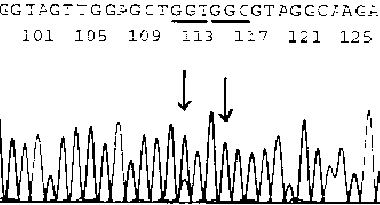Method for rapidly concentrating and extracting nucleic acid target cells from sample
A technology of cell nucleic acid and target cells, applied in the field of nucleic acid extraction, to achieve the effect of increasing the positive rate, increasing the concentration of nucleic acid, and good social effects
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0033] The dewaxing of embodiment 1 white sheet
[0034] Select several white sheets (2-15 sheets) depending on the size of the specimen and soak them in xylene for 8-12 hours to fully dissolve the paraffin; replace with new xylene to dissolve the paraffin and soak for 15-20 minutes; replace xylene again and soak for 15 minutes; Immediately soak the slices in absolute ethanol for more than 10 minutes to fully dissolve xylene; replace with new absolute ethanol, soak for 10 minutes, and then soak in 95% ethanol for 5 minutes; 80% ethanol for 5 minutes; 70% ethanol for 2 minutes; tap water for a while 2min, washed with distilled water for 2min, blotted dry, and then added dropwise 50uL of nucleic acid protection solution. Under the microscope, select the target cells on the HE stained sheet with a marker pen, and under the guidance of the HE sheet, also select the target cells with a marker pen at the same position on the white sheet, scrape off the target cells, enrich them, and...
Embodiment 2
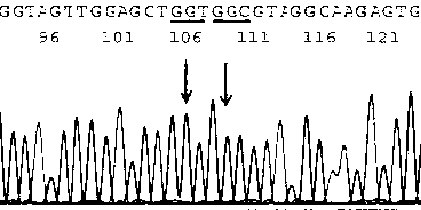
[0036] Example 2 Selection of target cells under the guidance of HE-stained slices of colorectal cancer (pathological number 082379)
[0037]HE-stained sections must be re-examined and diagnosed by pathologists or cytologists before nucleic acid extraction to confirm that they contain target cell components, and mark the location of target cells on the slide. Using the method of Example 1, extract the DNA template, and use the primers of the K-ras-2 gene (K-ras-2-U: -aggcctgctgaatttgactg-, K-ras-2-L: -catgattttggtcagagaaacc-;) for PCR amplification (QIAGEN HotStar DNA polymerase Kit10×amplification buffer 2uL, dNTP2U, upstream and downstream primers 10pmol each, template DNA 0.1~2ug Taq DNA polymerase 2.5u, Mg 2+ 1.5mmol / L add double distilled water to 20ul. PCR thermal cycle conditions: 95°C for 10min, 95°C for 40s, 60°C for 1min, 72°C for 45s, 30 cycles, 72°C for 45min, 4°C for incubation), the product was purified and sequenced (conventional method), the results are as f...
PUM
| Property | Measurement | Unit |
|---|---|---|
| melting point | aaaaa | aaaaa |
| thickness | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com