Method for assembly of nucleic acid sequence data
A nucleic acid sequence and nucleotide sequence technology, applied in sequence analysis, electronic digital data processing, special data processing applications, etc., can solve problems such as non-standardized raw data, valueless NGS sequencing machines, and differences in matching thresholds
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0127] Example 1 - Reference and de novo alignment of sequence reads to establish accurate repeat content of the AVPR1A gene
[0128] Since the repeat content (number of repeats) of the AVPR1A gene is associated with behavior, it has major health implications. Therefore, experimental assessments were performed based on reference and de novo alignments of sequence reads to establish the accurate repeat content of AVPR1A.
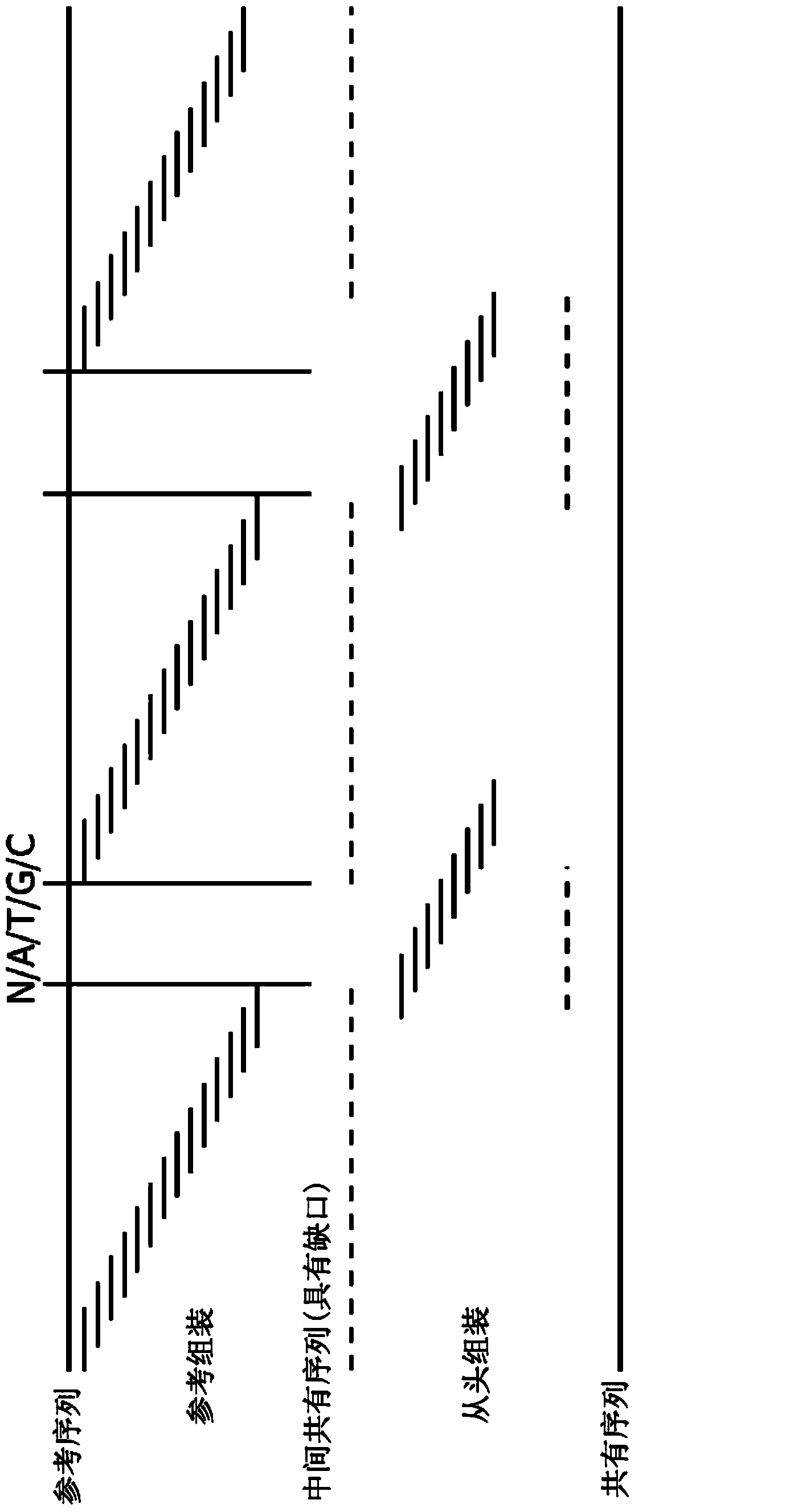
[0129] Reference alignments were used to map reads to genomic coordinates, while de novo alignments were used to determine the exact repeat content in the AVPR1A gene (see Figure 5 and 6 ).
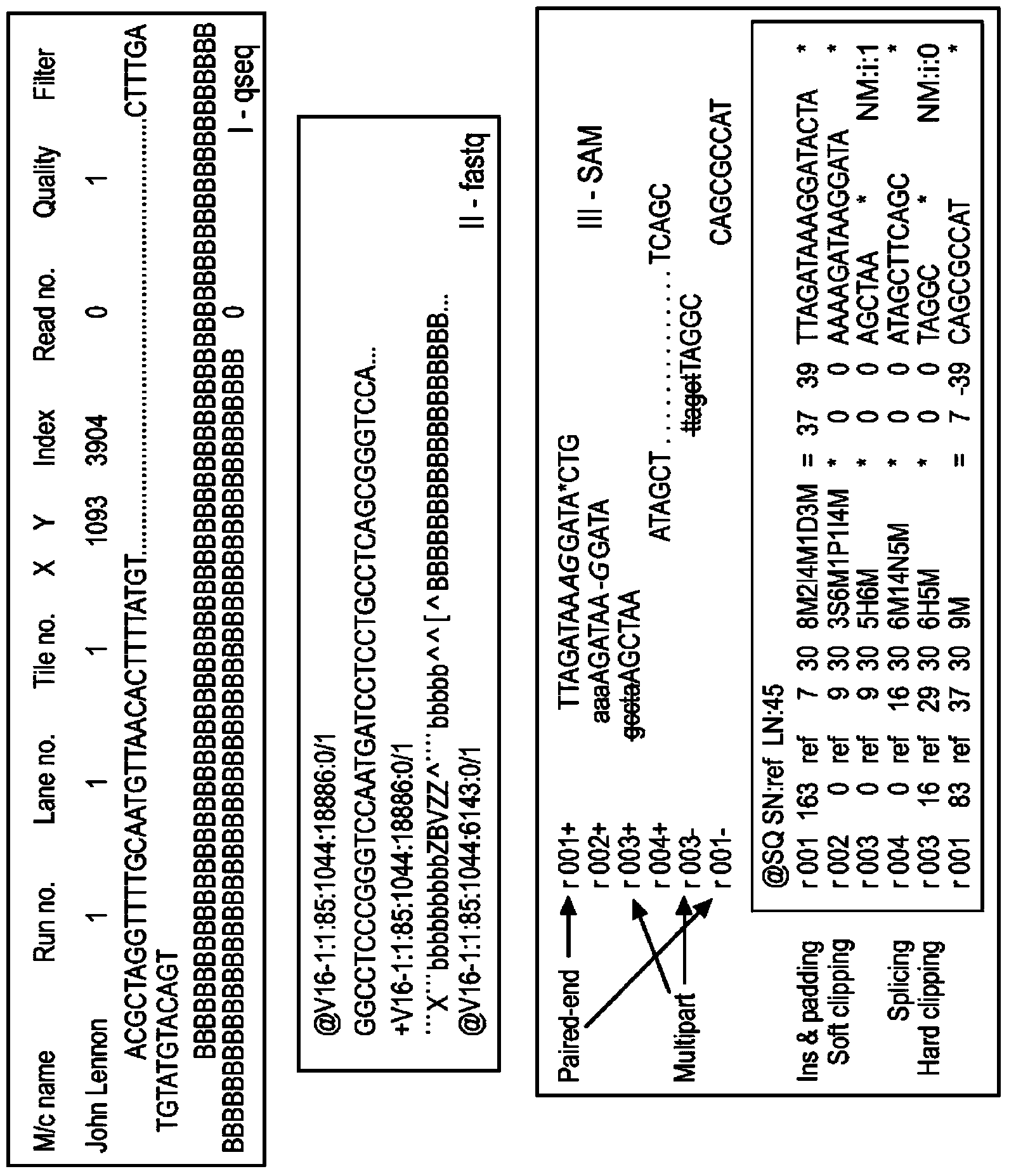
[0130] The Qseq files obtained from Illumina GAIIx were first converted into fastq format. These files were then aligned to the human reference (GRCh37) genome using the BWA aligner. Consensus sequences were constructed using the SAM output from the BWA alignment. We know that the RS3 polymorphism in the AVPR1 gene is actually highly polymorphic and associated with c...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com