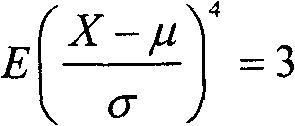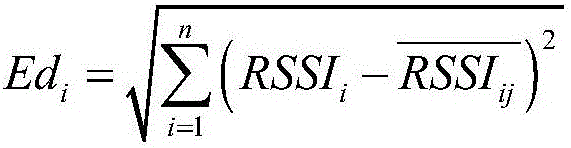Patents
Literature
372results about How to "Solve deviation" patented technology
Efficacy Topic
Property
Owner
Technical Advancement
Application Domain
Technology Topic
Technology Field Word
Patent Country/Region
Patent Type
Patent Status
Application Year
Inventor
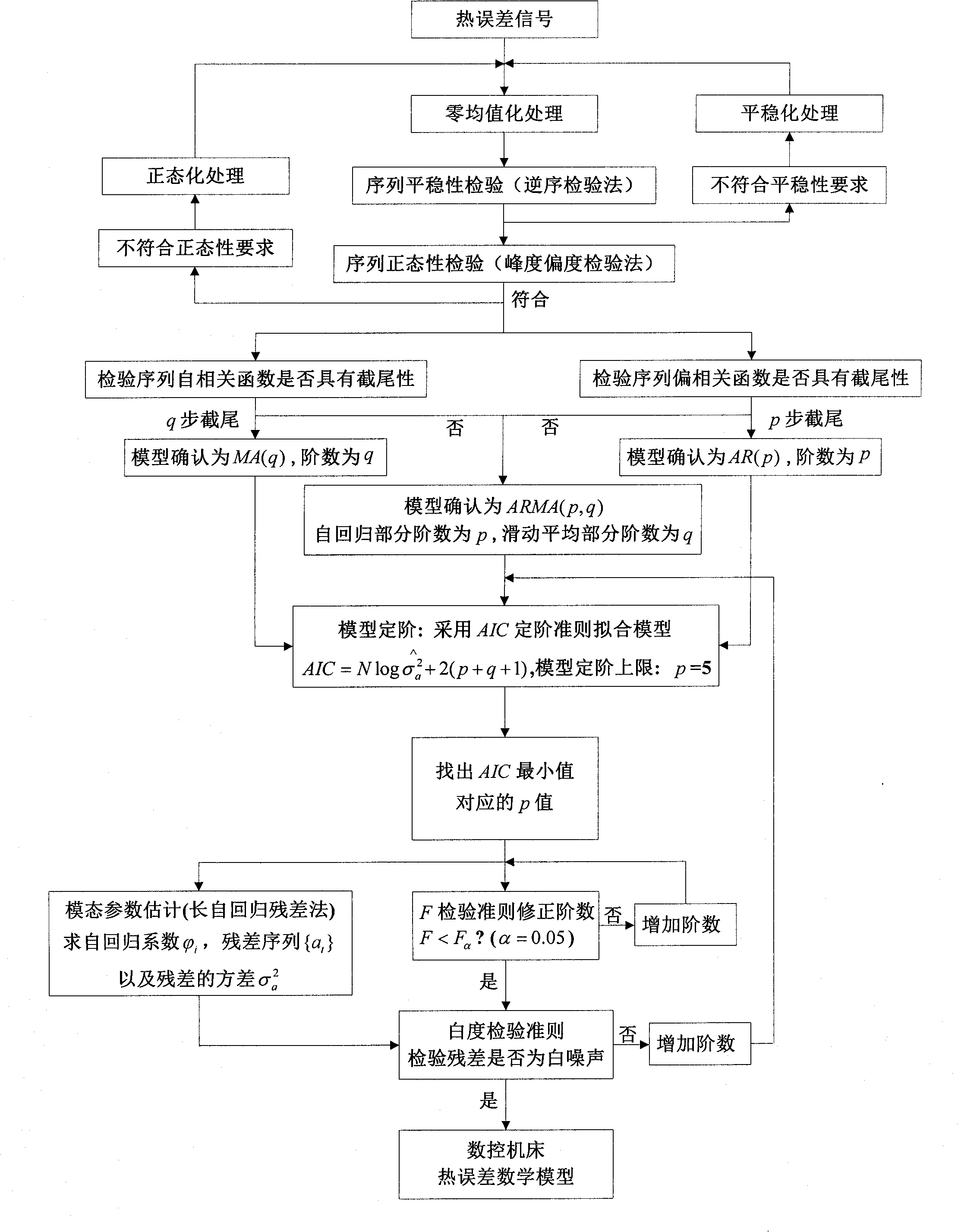
Numerical control machine tool thermal error real-time compensation modeling method based on time series algorithm
InactiveCN102736558AReduce hardware costsReduce complexityProgramme controlComputer controlNumerical controlMathematical model
The invention relates to a numerical control machine tool thermal error real-time compensation modeling method based on a time series algorithm, which belongs to the technical field of precision machining. The method comprises the steps of (1) carrying out data zero mean pretreatment, namely employing an inverted sequence test method and a kurtosis and skewness test method to judge the stationarity and the normality of the data; (2) using an autocorrelation function, a partial correlation function, and the censored results as judgment criteria to carry out the pattern recognition of a thermal error mathematical model; (3) employing a least square estimation method or a long autoregressive residual calculating method to realize the parameter estimation of the thermal error mathematical model; (4) determining the order of the thermal error mathematical model, namely employing a judgment method that combines an AIC order determination criterion, an F test order determination criterion, and a whiteness test order determination criterion to realize the order determination of the thermal error mathematical model; (5) and carrying out integration processing of synthesizing judgment conditions, namely constructing a complete forecasting mathematical model formula. The modeling method provided by the invention has the advantages that less hardware is required, the applicability is wide, and the established model has high prediction precision and reliability.
Owner:上海睿涛信息科技有限公司
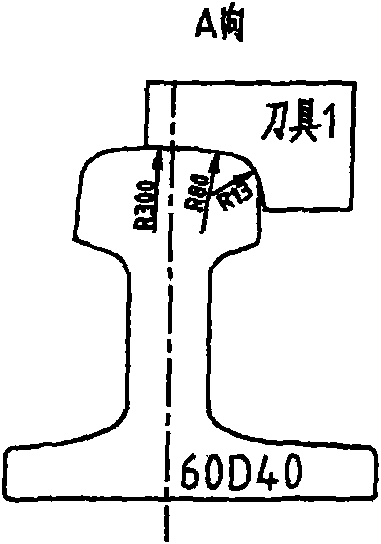
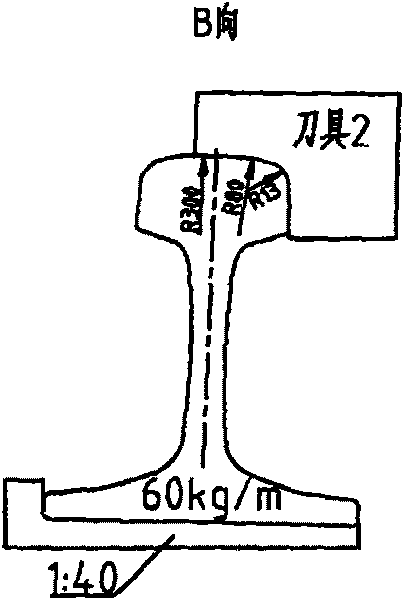
Processing technique for top surface of rail
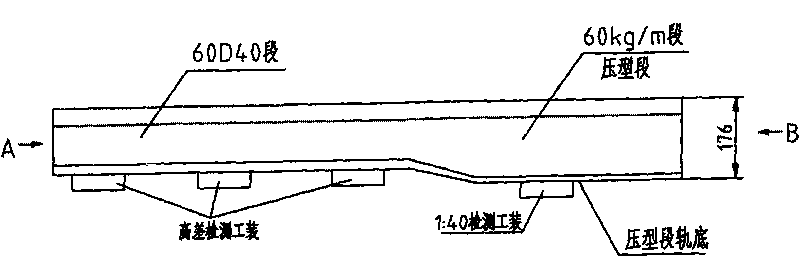
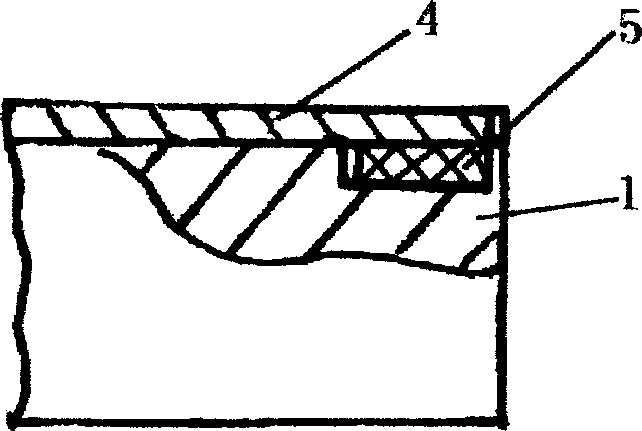
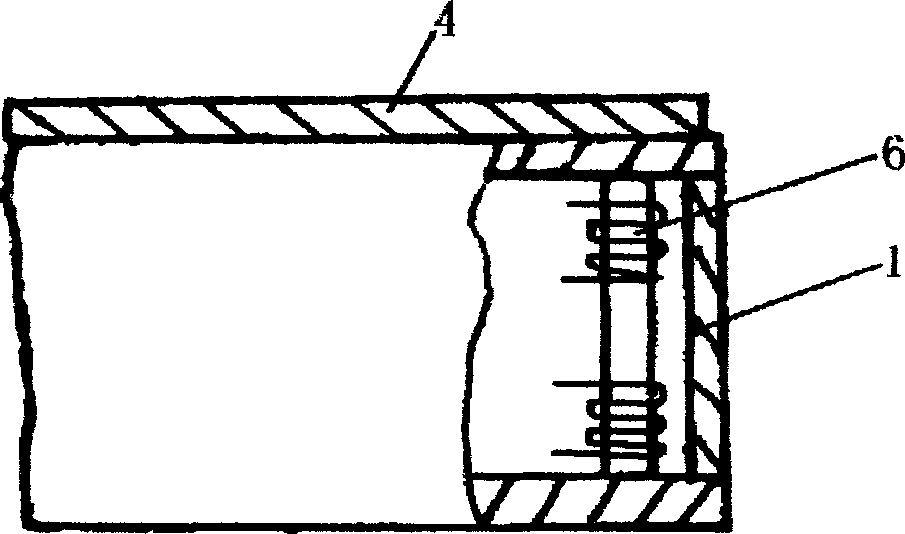
ActiveCN101716717AMake a smooth transitionSolve deviationMilling equipment detailsQuenchingMechanical engineering
The invention relates to a processing technique for the top surface of a rail, which is implemented by the following process flows: primary baiting; saw cutting; press molding; secondary baiting; aligning; milling a compressed section rail base; fine tuning; drilling; milling blot grooves of the rail base and rail web; milling a working edge and a non-working edge of a rail head; milling a rail top and derating; quenching; and leveling. The fine turning flow comprises the following steps of: firstly, leveling the rail base of the rail, ensuring that the flatness of the rail base is less than or equal to 0.5mm; secondly, obtaining the relative altitude difference of the rail base, a rail top surface and two sides of the rail head, detecting the straightness of the two sides of the compressed section rail head and the straightness of the rail top surface by using a leveling ruler, ensuring that an idle line between the two sides of the molded section rail head and raw materials of the rail is 1mm, and the top surface of the molded section rail is 1.5mm higher than the top surface of the raw materials of the rail; and finally, detecting the 1:40 inclination of a heal end and the altitude difference of the rail base by using an altitude difference testing tool and 1:40 inclined testing equipment. According to the processing technique, a novel copying tool is used for machining the rail top surface of the 60D40 rail, so the molded section and a transition section are in smooth transition with the raw materials; the fitting degree of the machined top surface of the 60D40 rail is more complete; and the machining accuracy of the products is guaranteed.
Owner:CHINA RAILWAY BAOJI BRIDGE GRP
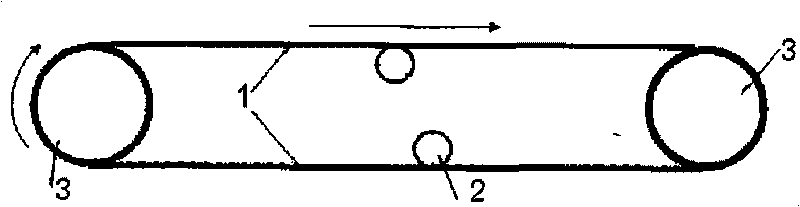
New correcting method and device of metal conveyor belt
The invention discloses new correcting method and device of a metal conveyor belt, belonging to the correction field of mechanical devices. In the invention, by adopting a correction mode of a permanent magnet or an electromagnet, the conveyor belt is limited in deviation by magnetic force or the deviated conveyor belt is pulled back to a normal track, i.e. grooves or convex edges are arranged at two ends of a driving roller of the metal conveyor belt, magnetic correcting devices with magnetism opposite to or same as that of the corresponding sides of the metal conveyor belt are arranged on two inner sides of the grooves or the convex edges, two sides of the metal conveyor belt are folded into folded edges with the depth corresponding to the depths of the grooves or the convex edges or small magnets are arranged at intervals on two sides of the metal conveyor belt so as to increase the magnetic action and strengthen the magnetic action effect. The invention is simple and feasible, effectively overcomes the defects of side deformation and cracking of the metal conveyor belt caused by the deviation in the background art and ensures normal production.
Owner:CHINA CHEM ENG SECOND CONSTR
Method for assembly of nucleic acid sequence data
InactiveCN103797486ASolve deviationIncrease sequence precisionSequence analysisSpecial data processing applicationsNucleotideNucleic acid sequence
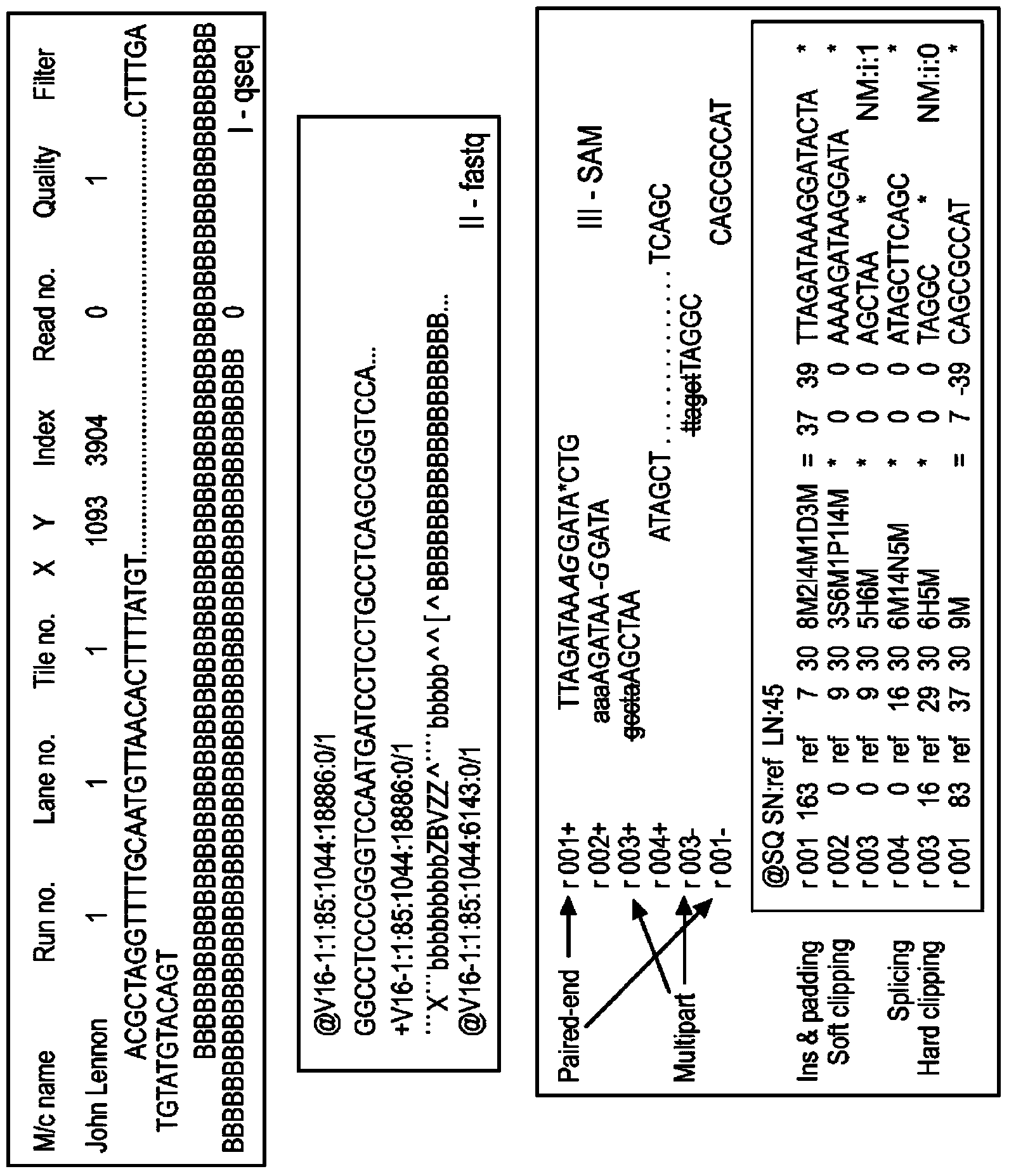
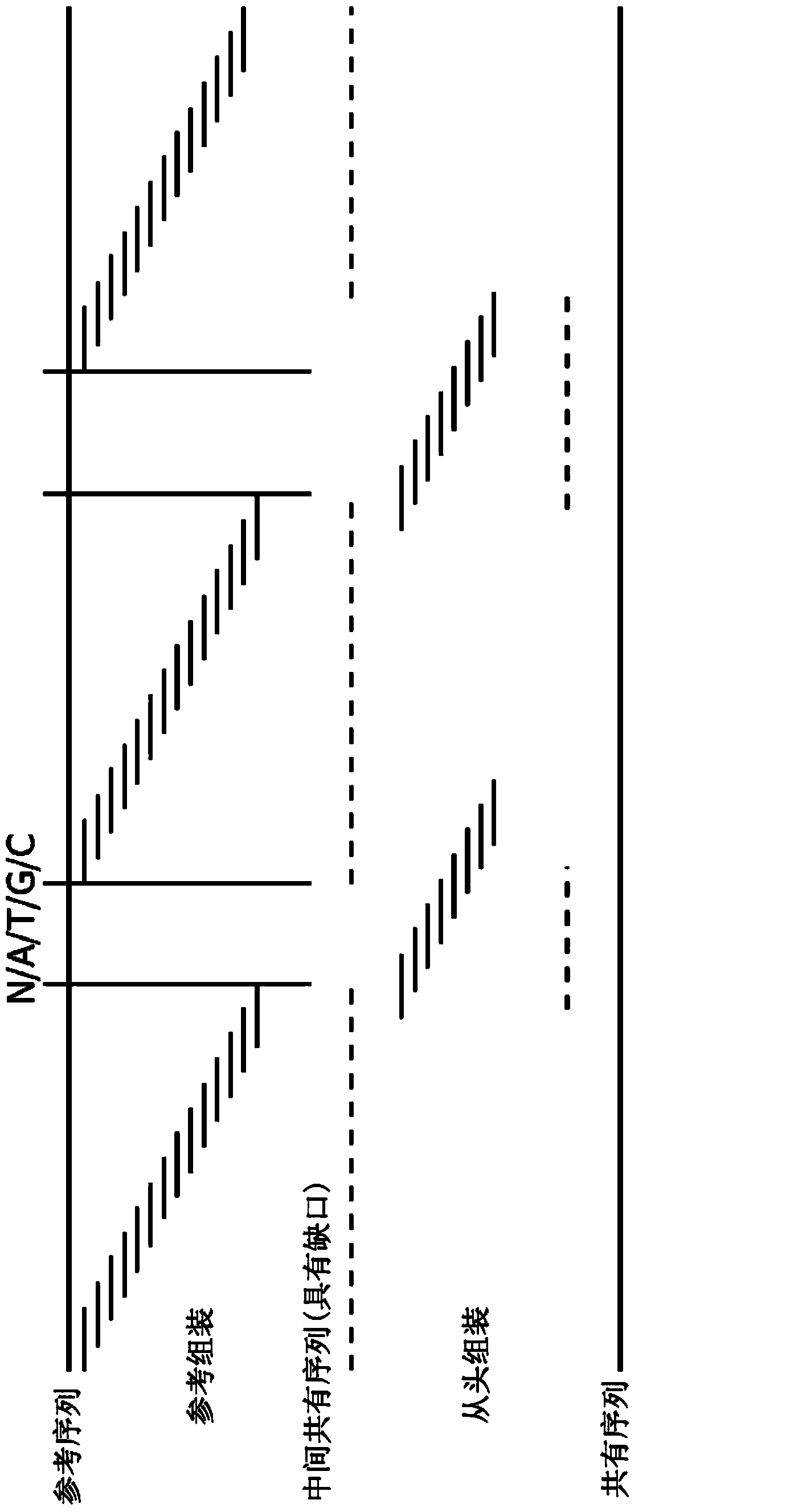
The present invention relates to a method for assembly of nucleic acid sequence data comprising nucleic acid fragment reads into (a) contiguous nucleotide sequence segment(s). The method comprises steps of: (a) obtaining a plurality of nucleic acid sequence data from a plurality of nucleic acid fragment reads; (b) aligning the plurality of nucleic acid sequence data to a reference sequence; (c) detecting one or more gaps or regions of non-assembly, or non-matching with the reference sequence in the alignment output of the step (b); (d) performing de novo sequence assembly of nucleic acid sequence data mapping to the gaps or regions of non-assembly; and (e) combining the alignment output of the step (b) and the assembly output of the step (d) in order to obtain (a) contiguous nucleotide sequence segment(s).The present invention further relates to a method, wherein the detection of gaps or regions of non-assembly is performed by implementing a base quality, coverage, complexity of the surrounding region, or length of mismatch filter or threshold. Also envisaged is the masking out of nucleic acid sequence data relating to known polymorphisms, disease related mutations or modifications, repeats, low map ability regions, CPG islands, or regions with certain biophysical features. In addition, a corresponding program element or computer program for assembly of nucleic the sequence data and a sequence assembly system for transforming the nucleic acid sequence data comprising nucleic acid fragment reads into (a) contiguous nucleotide sequence segment(s) are provided.
Owner:KONINKLJIJKE PHILIPS NV
RSSI corrected wireless sensor network positioning algorithm of self-adaptive environment
InactiveCN105813020AHigh precisionSolve deviationNetwork topologiesUsing reradiationSmall sampleComputation complexity
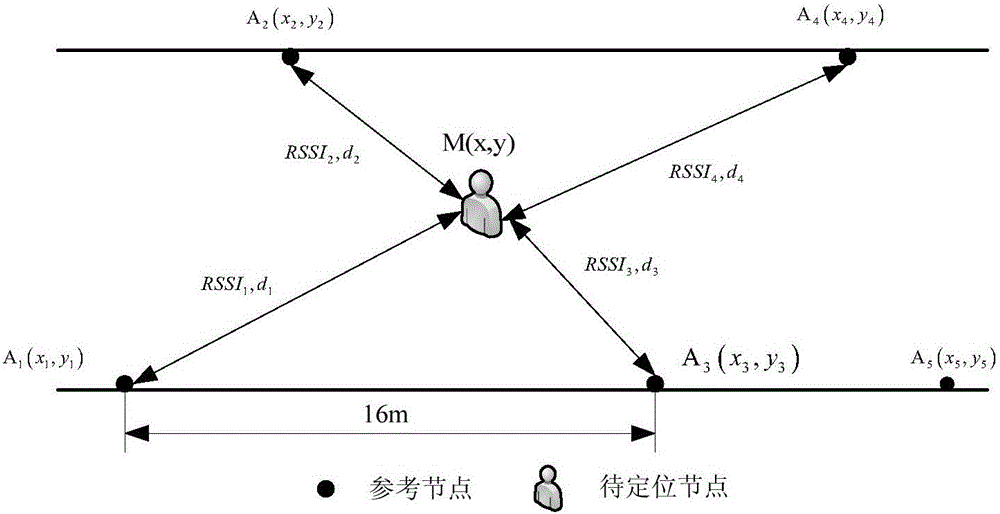
The invention discloses a RSSI corrected wireless sensor network positioning algorithm of a self-adaptive environment. The method comprises constructing an offline fingerprint database using a confidence interval thought to realize the aim of estimating the total true value using the small sample measurement value. In the process of promoting the precision, the K value in the K-NNSS algorithm needs to weigh between the algorithm precision and the computation complexity. Besides the fingerprint positioning, the RSSI value acquired through the filtering of a Gaussian model is specifically used, and the distance relation between a unknown node and a beacon and the distance relation between the beacons are combined so as to correct the distance between the unknown node and the beacon, the accuracy of the maximum likelihood of location solution is improved; the corrected distance relation between the unknown node and the beacon is used for constructing a subsequent centroid positioning weighting coefficient, thereby reflecting the positioning influence of each beacon to the unknown node to promote the precision of the algorithm.
Owner:HEFEI UNIV OF TECH
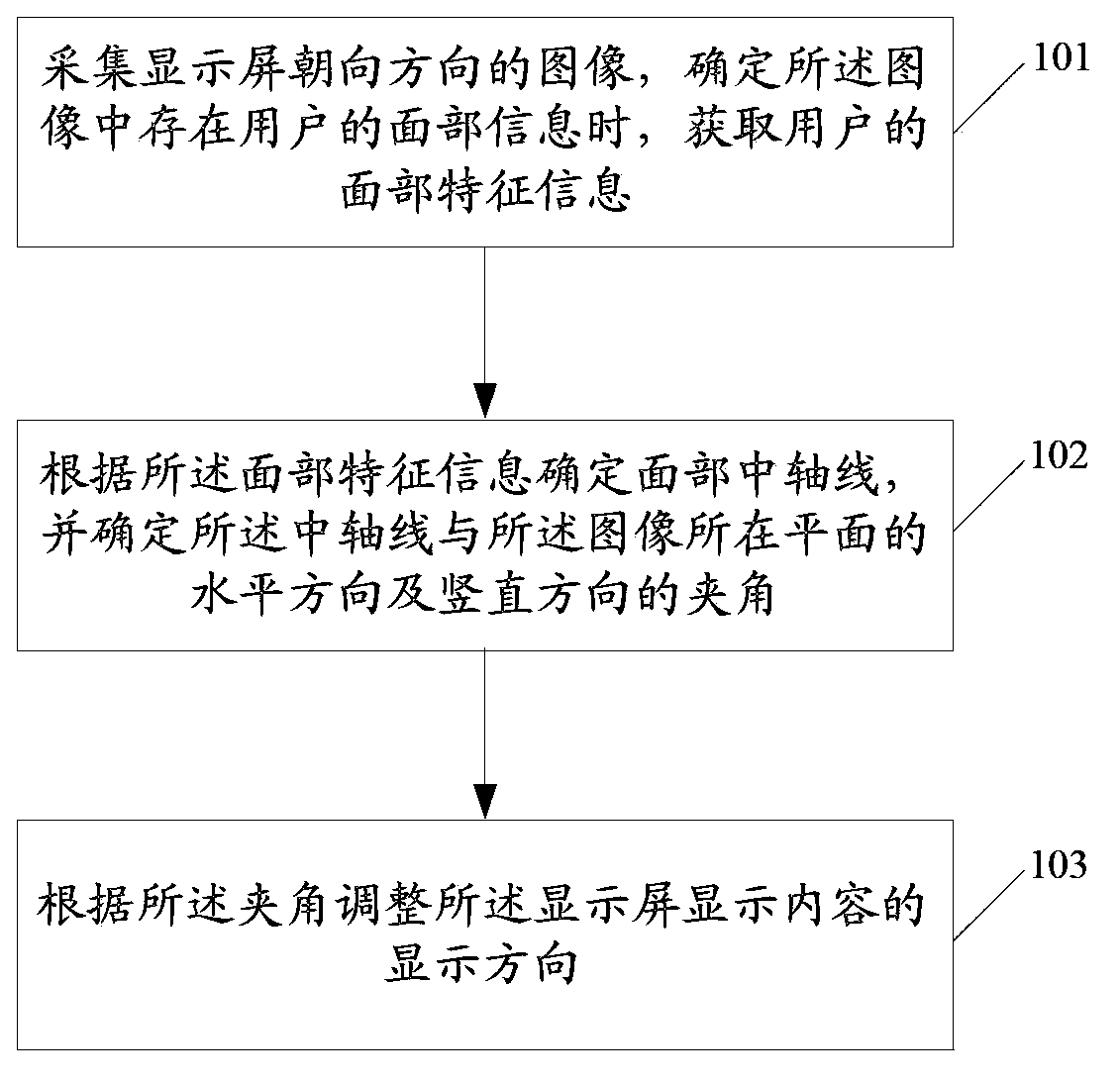
Adjusting method and device for screen displaying direction
InactiveCN104122983AImprove experienceImprove display efficiencyInput/output for user-computer interactionGraph readingFree stateComputer vision
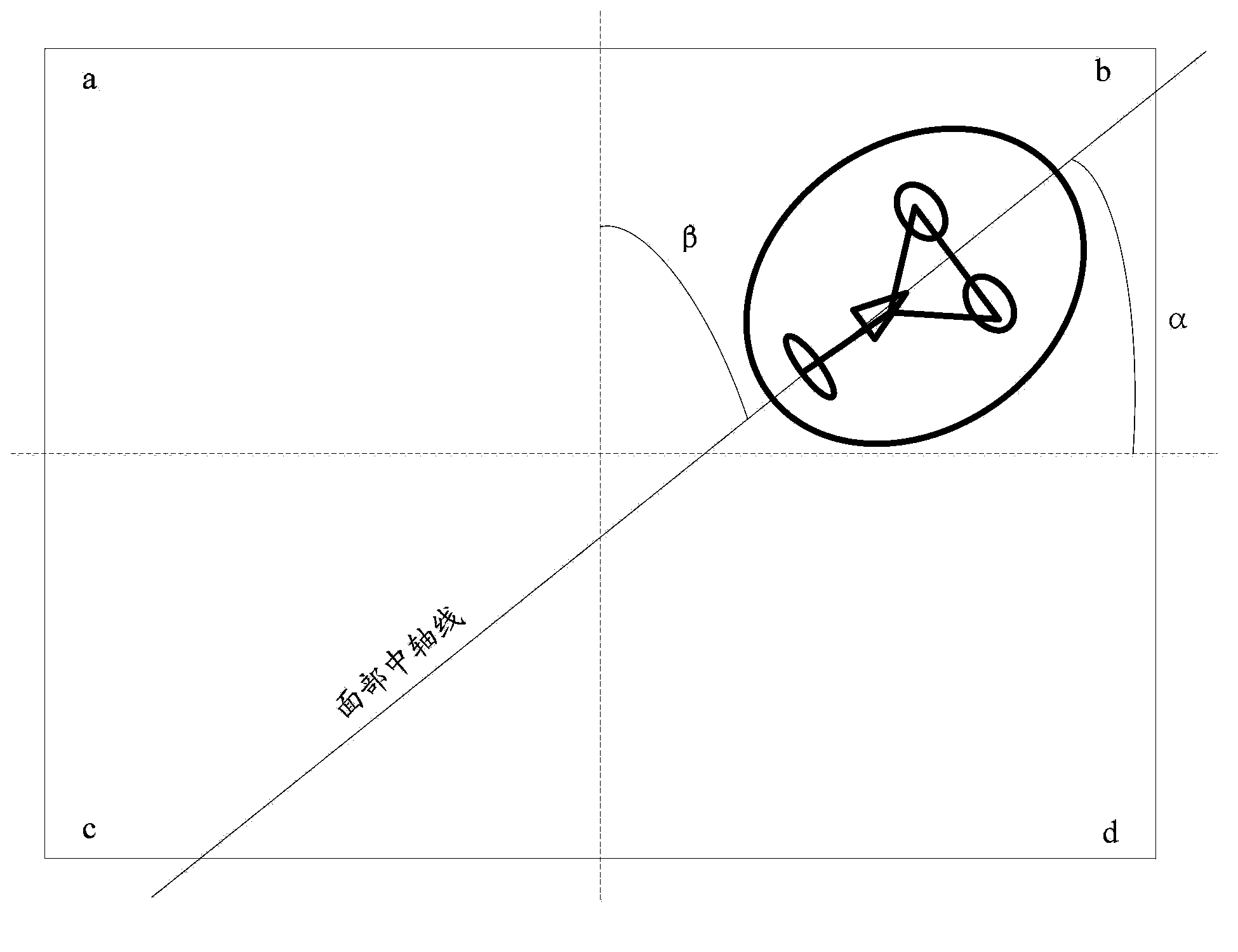
The invention discloses an adjusting method for the screen displaying direction. An image in the facing direction of a display screen is collected, and when it is determined that face information of a user exists in the image, face characteristic information of the user is obtained; according to the face characteristic information, the central axis of the face is determined, and the included angles between the central axis and the horizontal direction of the plane where the image is located as well as between the central axis and the vertical direction of the plane where the image is located are determined; according to the included angles, the displaying direction of display content of the display screen is adjusted. The invention further discloses an adjusting device for the screen displaying direction. According to the scheme, the problem that when a terminal is held in an unconventional gesture or the terminal is in a gravity-free state, the screen displaying direction of the terminal deviates can be solved, meanwhile, the displaying efficiency of the terminal is improved, and better application experience is brought to the user.
Owner:CHINA MOBILE COMM GRP CO LTD
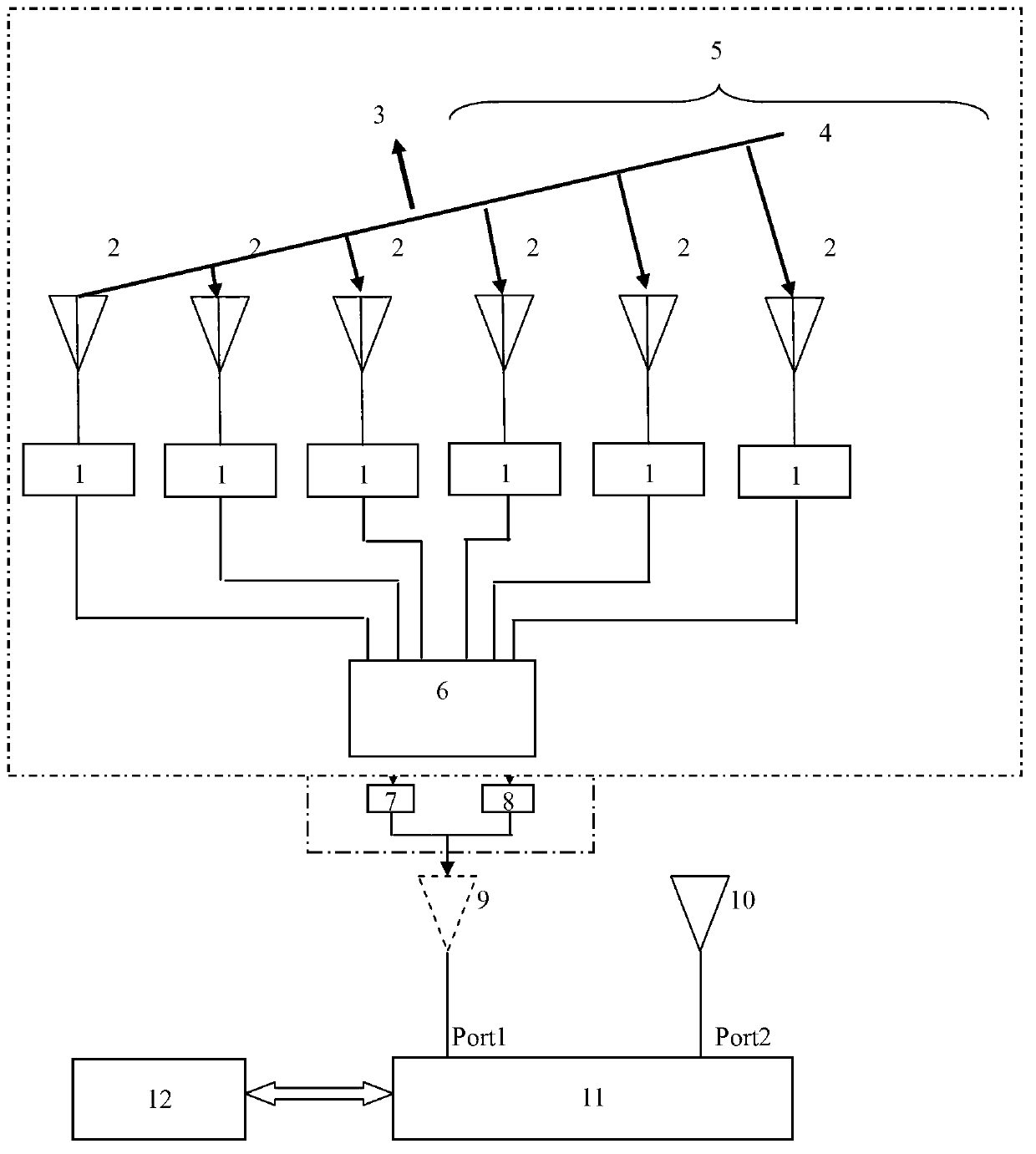
Phased array antenna equivalent isolation degree testing method
ActiveCN103217589AHigh precisionImprove test efficiencyAntenna radiation diagramsArbitrary unitAntenna gain
The invention discloses a plane antenna engineering equivalent testing method and particularly relates to a phased array antenna equivalent isolation degree testing method. The array antenna isolation degree is calculated in the first place, an arbitrary unit array element of a phased array antenna is subsequently chosen as a testing reference point, and then a gain correction coefficient of the array antenna is calculated. The plane antenna engineering equivalent testing method can successfully solve engineering test deviation due to uncertainty of gain of an antenna main lobe and the antenna, wherein the uncertainty of the gain of the antenna main lobe and the antenna is caused by variation of array antenna phase synthesis. By introducing correction factors such as the gain after phase synthesis, an antenna array angle and beam width deviation, the phased array antenna is changed to be an equivalent unit antenna. The phased array antenna equivalent isolation degree testing method is suitable for isolation degree engineering tests between the phased array antenna and a unit antenna and between the array antennas and can be popularized and applied to array antenna isolation degree engineering tests on all sorts of weapon equipment platforms.
Owner:SHAANXI AIRCRAFT CORPORATION
Load correction method applied to undercarriage loading
ActiveCN105083587AGuaranteed to be verticalReduce load errorAircraft components testingControl theoryReliability engineering
The invention discloses a load correction method applied to undercarriage loading. The method comprises the steps of 101, installing a loading device on an undercarriage; 201, acquiring corresponding test data between the offset, in the heading direction of the undercarriage, at the position where the loading device is connected with the undercarriage and a load multiple times through testing; 301, fitting the test data obtained in the step 201 into a formula by means of the least square method; 401, obtaining the offset, in the heading direction of the undercarriage, at the position where the loading device is connected with the undercarriage according to a preset load by means of the formula obtained in the step 301; 501, adjusting the loading device so as to counteract the offset. In this way, load error generated by an undercarriage loading system is effectively reduced so that the test load error can meet the control requirement, and then the problem that offset exists in the prior art is solved.
Owner:XIAN AIRCRAFT DESIGN INST OF AVIATION IND OF CHINA
Optimizing design method of nanometer technical metal layer map
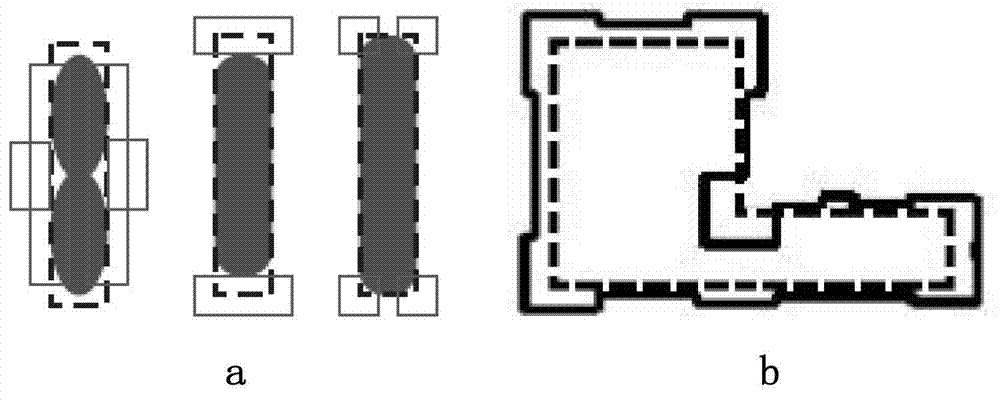
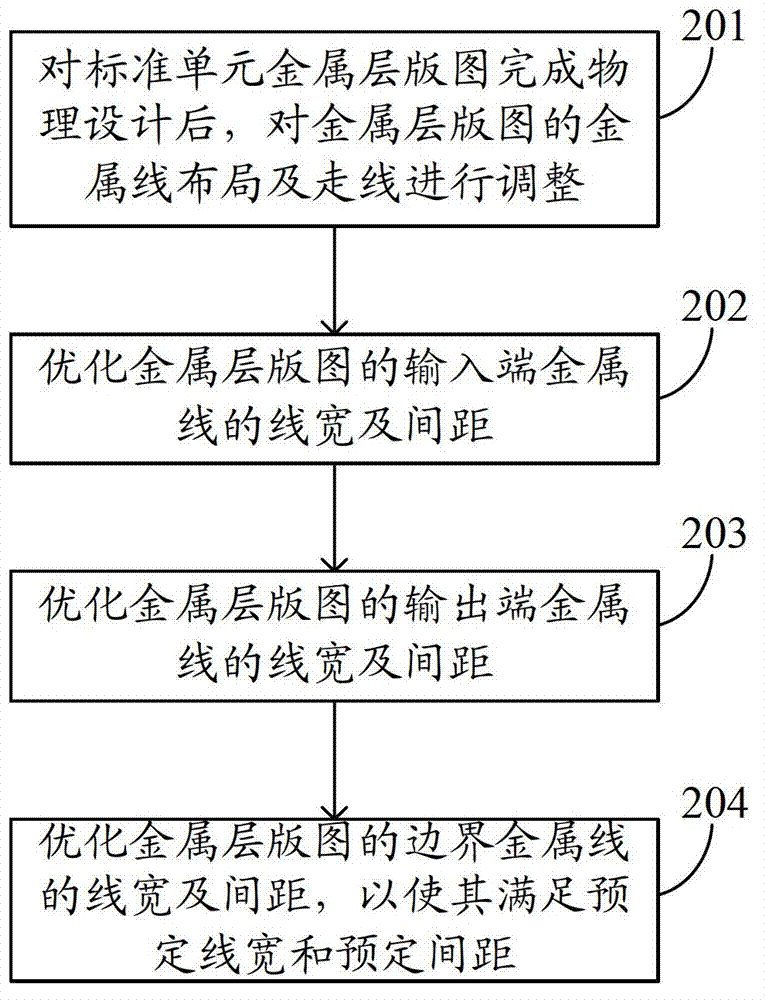
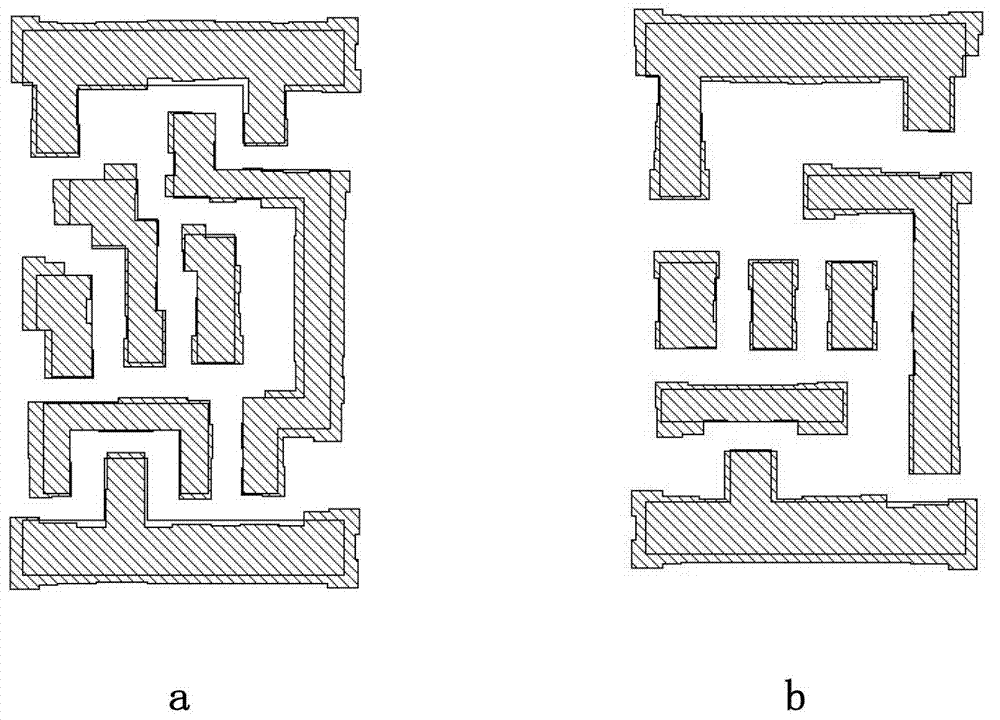
InactiveCN102855360AReduce cornersUniform density distributionSpecial data processing applicationsDensity distributionOptoelectronics
The embodiment of the invention provides an optimizing design method of a nanometer technical metal layer map. The method comprises the steps as follows: physically designing a standard unit metal layer map, and then adjusting the arrangement and direction of metal lines of the metal layer map; optimizing the width and spacing of the metal lines at the input end of the metal layer map; optimizing the width and spacing of the metal lines at the output end of the metal layer map; and optimizing the width and spacing of the metal lines at the boundary of the metal layer map, so as to meet the preset width and the preset spacing of the metal lines. With the adoption of the optimizing method provided by the embodiment of the invention, the turning in the metal layer map is reduced, and the metal layer map can be uniform in density distribution; the optics proximity correction and modification area required by the metal layer map can be optimized, the data volume in the optics proximity correction and modification and the modification time can be reduced, the manufacturability of a chip is improved, and the manufacturing cost of the chip is saved.
Owner:INST OF MICROELECTRONICS CHINESE ACAD OF SCI
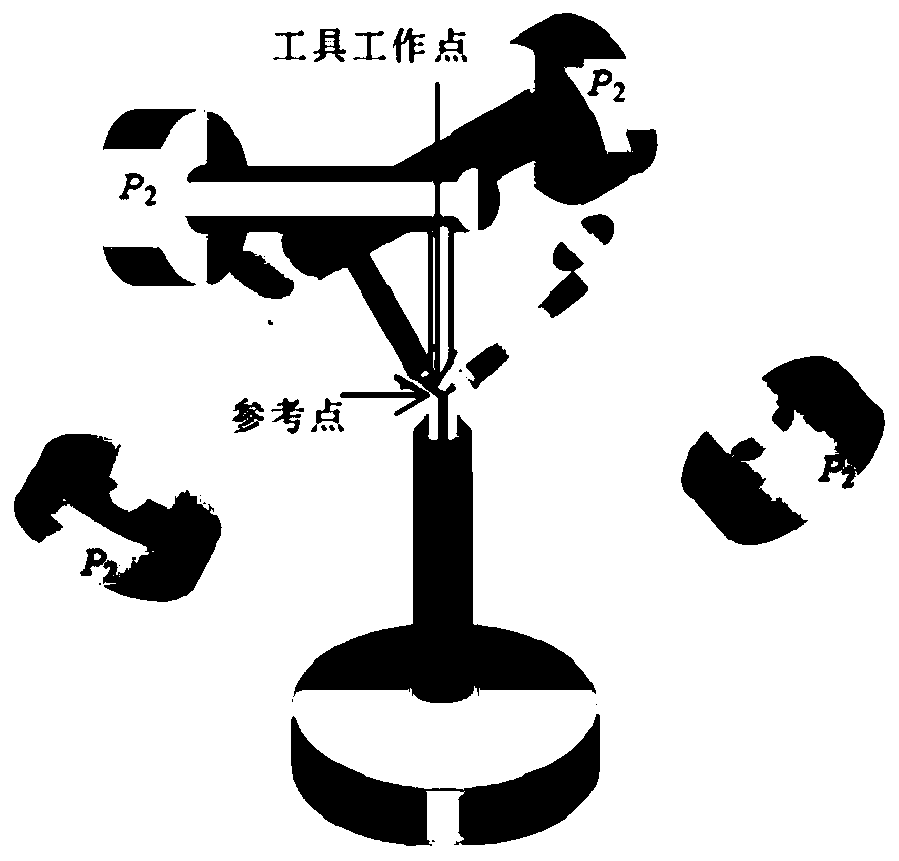
Tool center point calibration method and system for robot vision tool
InactiveCN109900207AEliminate manual teaching errorsEasy to operateUsing optical meansVisual toolComputer vision
The present invention belongs to the technological field of robot vision tools and provides a tool center point calibration method and system for a robot vision tool. The method includes the followingsteps that: a visual space coordinate system corresponding to a vision system is constructed according to the visual position information of a calibration reference plate collected by the vision system, wherein the calibration reference plate is capable of indicating a target object coordinate system; the calibration coordinate information of the calibration reference plate relative to a robot base coordinate system is obtained; and the tool center point of the robot vision tool is calibrated according to the calibration coordinate information and visual coordinate information corresponding to the visual position information, wherein the visual coordinate information is located in the visual space coordinate system. Therefore, multi-pose manual demonstration for a robot is not required; the automatic calibration process of the tool center point of the robot vision tool can be realized just by means of the calibration reference plate; and therefore, operation is convenient, and high precision can be realized.
Owner:精诚工科汽车系统有限公司
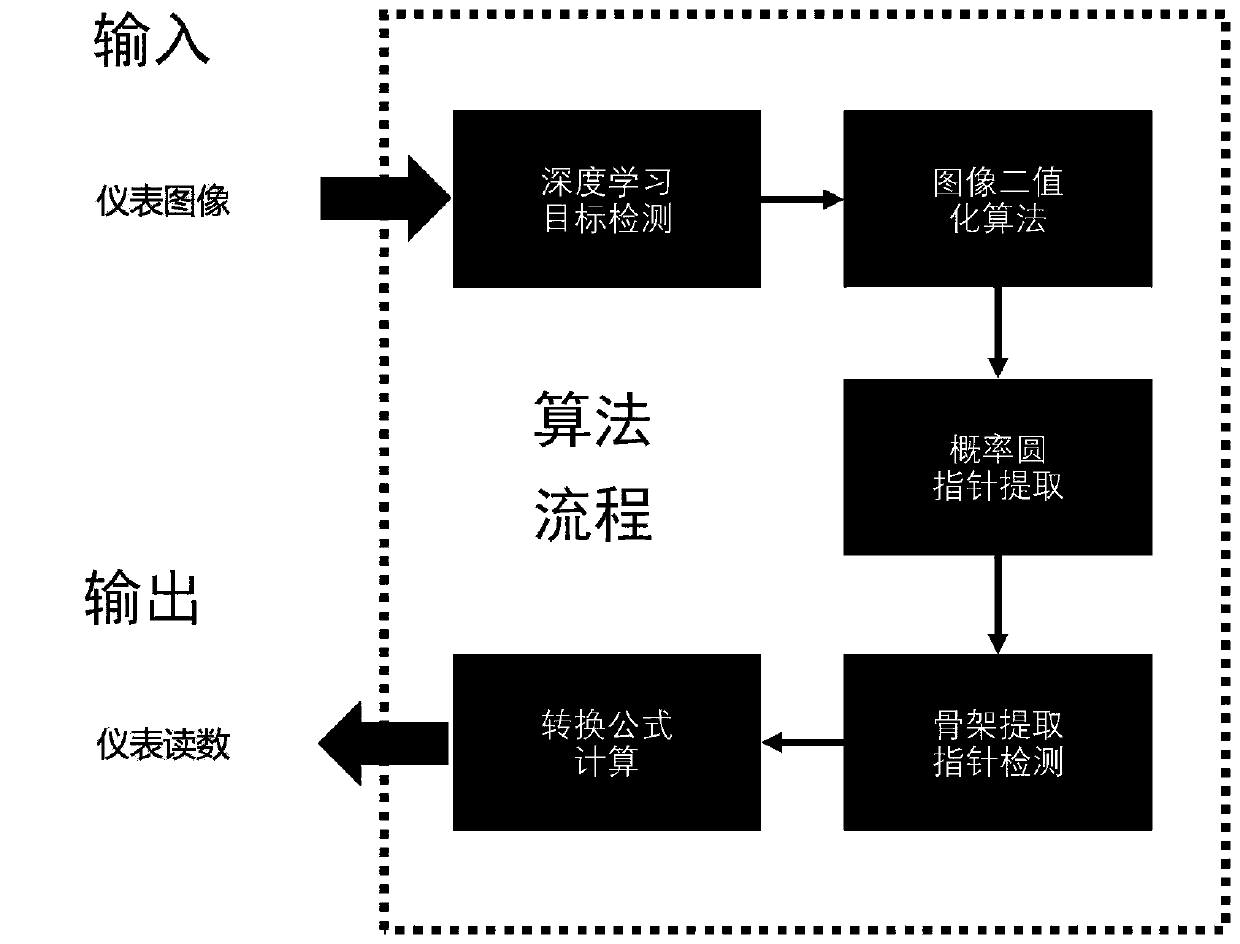
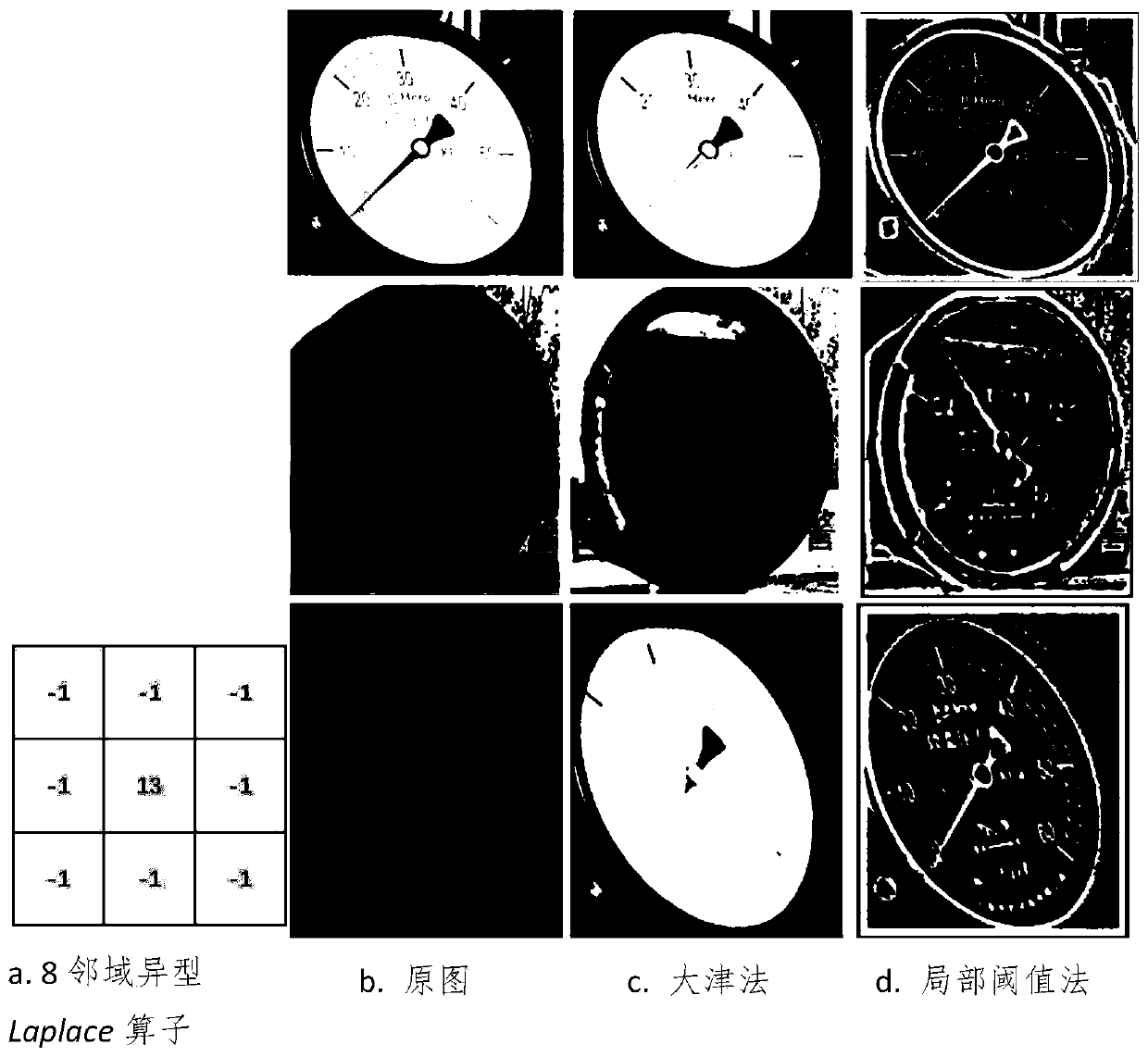
Pointer instrument detection and reading identification method based on mobile robot
ActiveCN110807355AImprove detection accuracyImprove the detection accuracy of small-scale targetsImage enhancementImage analysisEngineeringContrast enhancement
The invention relates to a pointer instrument detection and reading identification method based on a mobile robot. The method comprises the following steps: obtaining a deep neural network detection model M for a pointer instrument; the mobile robot moves to a designated place to obtain an original environment image containing the instrument equipment at present in a mode that the mobile robot carries a camera; S serves as system input and is transmitted to a deep neural network model M, whether an instrument exists in S or not is detected, the position of the instrument is framed out, an image in a frame is intercepted and subjected to height setting processing, the length-width ratio is not changed, and the processing result is represented by J; contrast enhancement is performed on the image, and a processing result is represented by E; local adaptive threshold segmentation is performed on the E to obtain a reverse binary image B; pointer extraction processing is performed based on aprobability circle; a central straight line L of the pointer part is extracted as pointing information of the instrument pointer; and a coordinate system is established based on a probability circlecenter projection algorithm and reading by an angle method.
Owner:TIANJIN UNIV
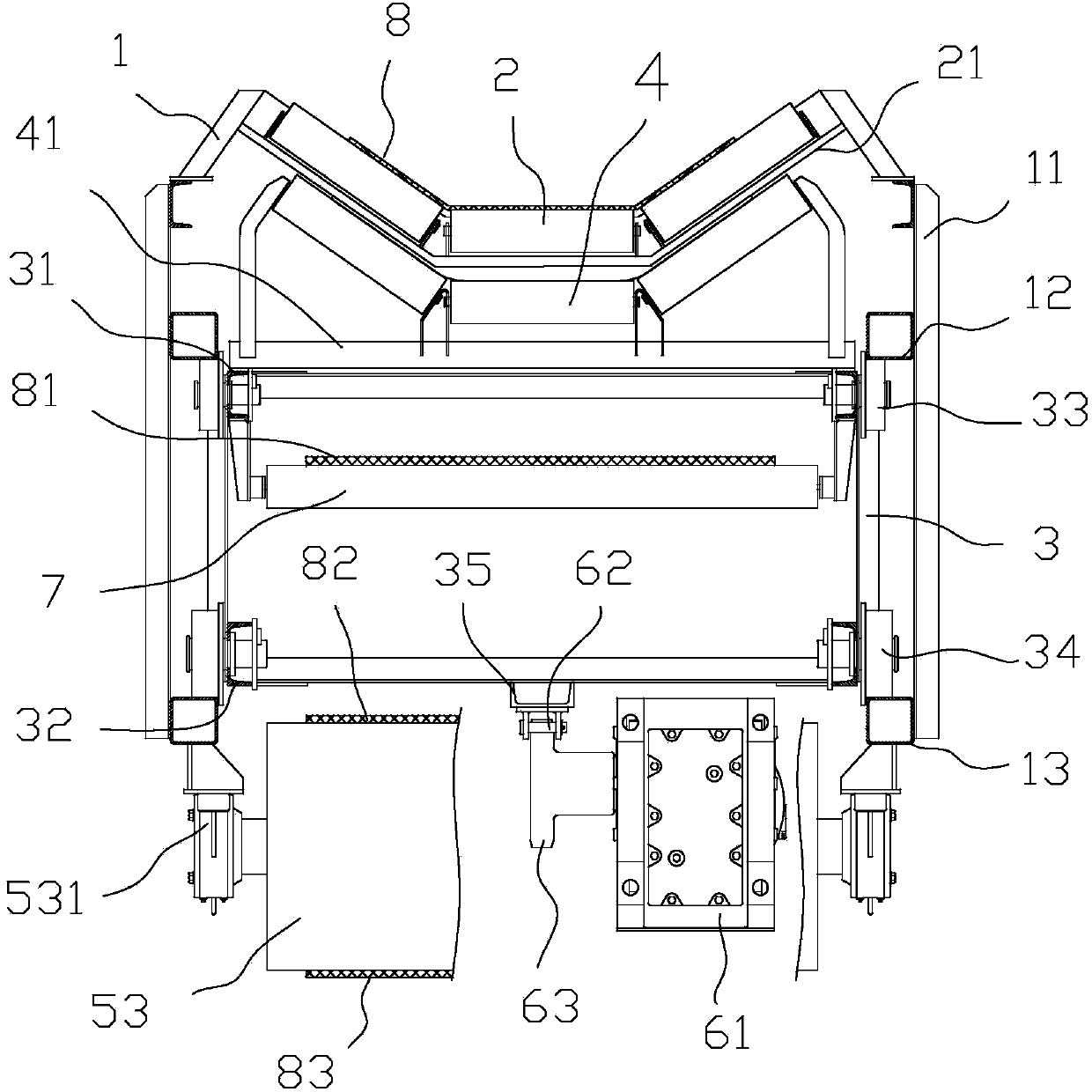
Fully-enclosed dustproof energy-saving anti-deviation belt conveyer
The invention provides a fully-enclosed dustproof energy-saving anti-deviation belt conveyer, and relates to energy-saving sealing improvement of an upper-layer operation carrier roller assembly of the belt conveyer and whole-course enclosed dust prevention of a receiving section, a conveying section and an unloading section of the conveyer. The belt conveyer has numerous energy consumption and easily produces dust in operation, while the existing common dust control mode can not realize better closed effect, service efficiency of dust-collecting equipment is low and belt wear is severe; and a great amount of dust still raises in the conveying section and the unloading section, so the dust problem can not be solved fundamentally. In the invention, multiple sealing assemblies are mounted with a V-shaped sealed energy-saving roller support as a fixed pivot, and a flexible material is taken as a main material for shielding dust and in seamless connection with casings of the receiving section and the unloading section casing and the sealing assemblies of a running section of the conveyer to form a membrane structural casing with multiple closed isolation chambers, thus realizing sealed dust prevention of the whole conveyer, solving the problems of belt deviation and belt wear caused by a baffle and enhancing energy-saving performance of a conveying system.
Owner:李超
Method for maintaining position of thin belt
InactiveCN1413894ASlow down the speed of deviationControl deviationNon-mechanical conveyorsRollersEngineeringMagnet
An offset correcting method for thin driving belt has two approaches. One approach is used of magnetic attraction, that is, the permanent-magnet material is installed on the driving roller or the electromagnetic coil is installed in the driving roller and magnetic thin driving belt is used. Another approach is to arrange two pressing friction rollers at both ends of driving roller above the driving belt and regulating their pressure and tension can correct the offset of thin driving belt.
Owner:BEIJING INSTITUTE OF CLOTHING TECHNOLOGY
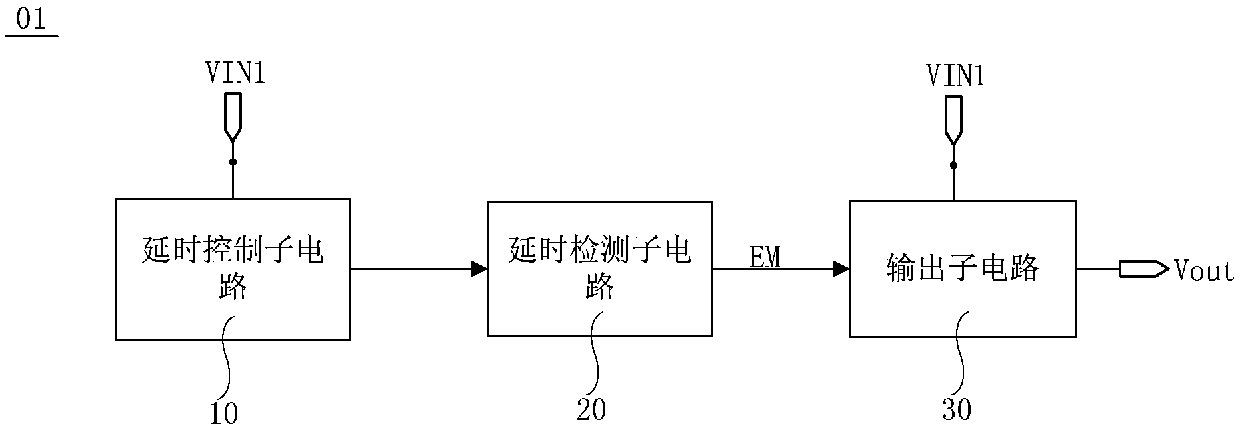
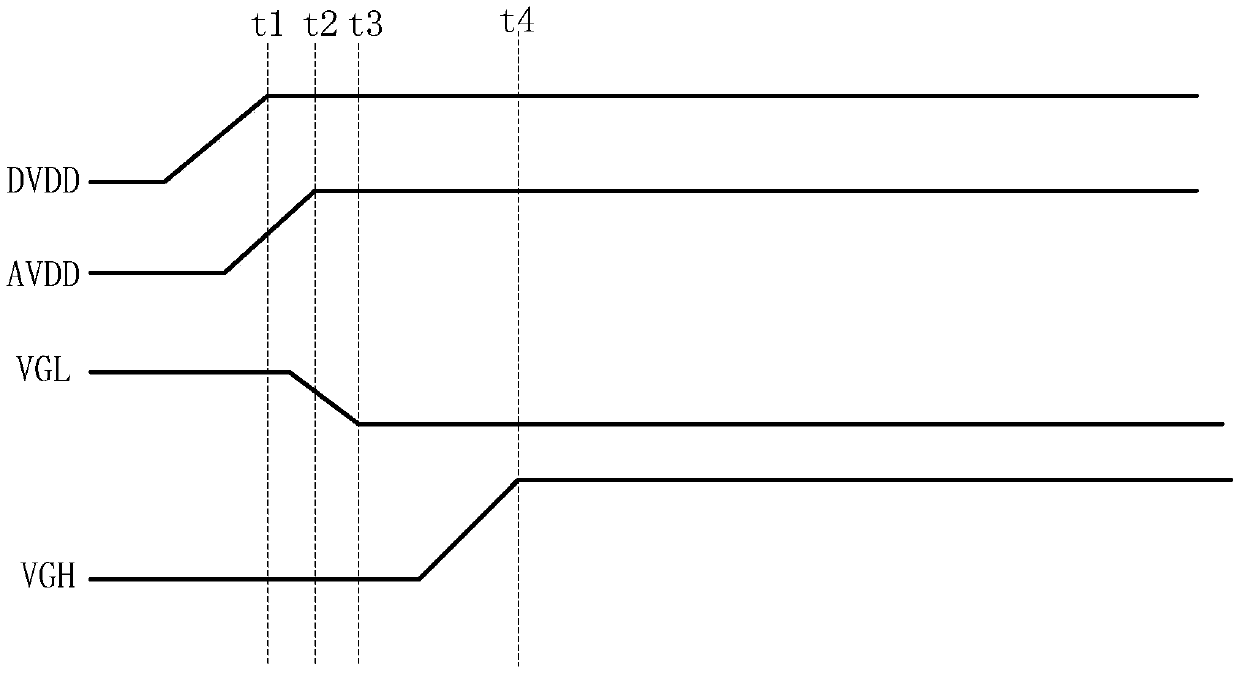
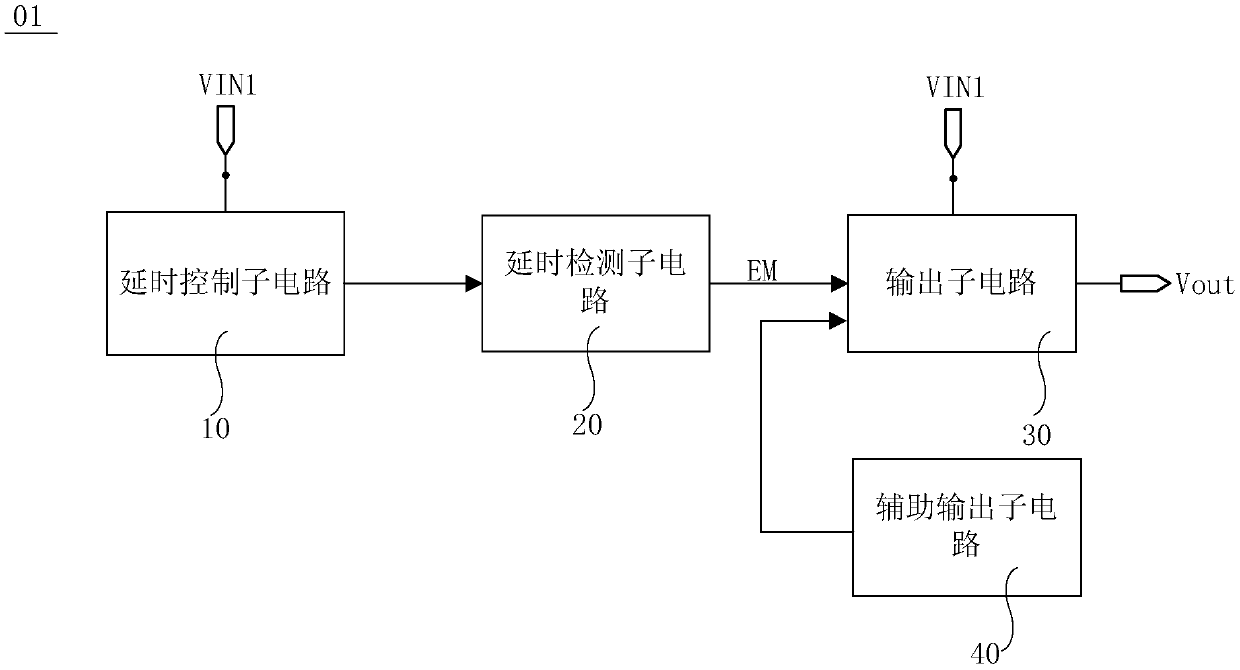
Power supply time sequence control circuit and control method, display driving circuit and display device
ActiveCN110544452ASolve deviationImprove stabilityStatic indicating devicesTime delaysDisplay device
The embodiment of the invention provides a power supply time sequence control circuit, a control method, a display driving circuit and a display device, relates to the technical field of display, which are used for solving the problem of deviation of a power supply time sequence caused by software programming control of the power supply time sequence. The power supply time sequence control circuitcomprises a time delay control sub-circuit, a time delay detection sub-circuit and an output sub-circuit. The delay control sub-circuit is used for delaying the first voltage output by the first input voltage end for a preset time and then outputting the delayed first voltage. The delay detection sub-circuit is used for sending a trigger signal to the output sub-circuit when detecting that the first voltage is received after the preset time. The output sub-circuit is further used for being in an on state according to the trigger signal and outputting the first voltage of the first input voltage end to the signal output end.
Owner:BOE TECH GRP CO LTD +1
Testing method for heat locating errors of linear shaft of vertical machining center
InactiveCN106975983AImprove accuracyAvoid influenceMeasurement/indication equipmentsGeometric errorMeasurement point
The invention discloses a testing method for the thermal positioning error of a linear axis of a vertical machining center. This method first measures the positioning error of each measuring point of the linear axis of the vertical machining center at the initial moment, that is, the geometric error, and then measures the positioning error of each measuring point of the linear axis of the vertical machining center at any time, and the conditions remain unchanged during the two measurements , and finally make a difference between the two data to obtain the thermal positioning error of each measurement point of the linear axis of the vertical machining center at any time. In this way, the influence of the geometric errors of the various components of the machine tool on the thermal positioning error measurement is avoided, and the accuracy of the measurement results is improved. This method can quickly and efficiently complete the on-machine test requirements of the thermal positioning error of the linear axis, which not only avoids the influence of the geometric errors of the various components of the machine tool on the measurement of the thermal positioning error, but also avoids the use of displacement sensors or standard parts as well as clamping, finding The deviation problem caused by positive and fixed improves the accuracy of the measurement results.
Owner:HEBEI UNIV OF TECH
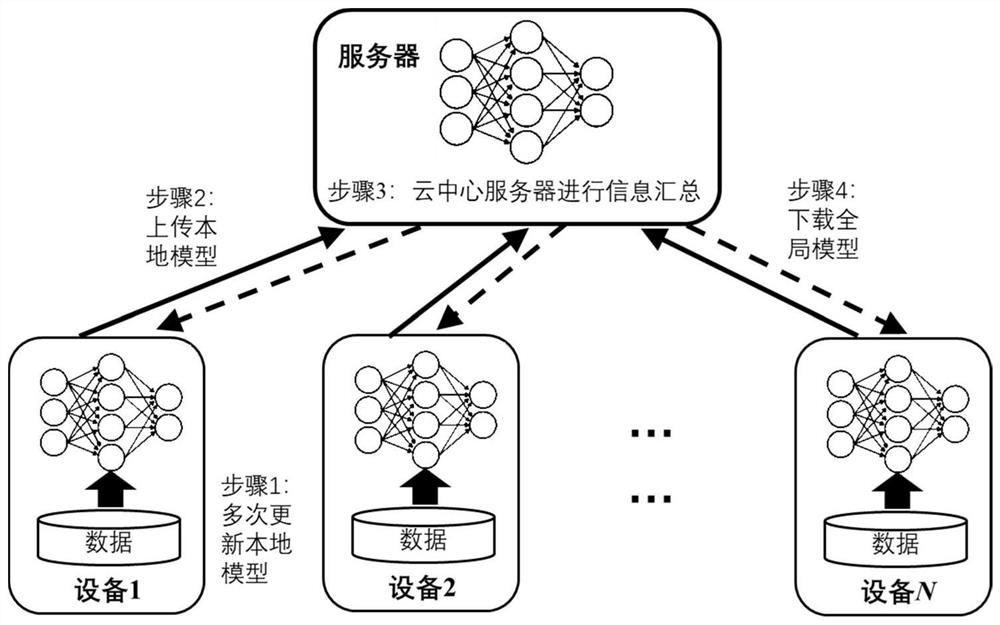
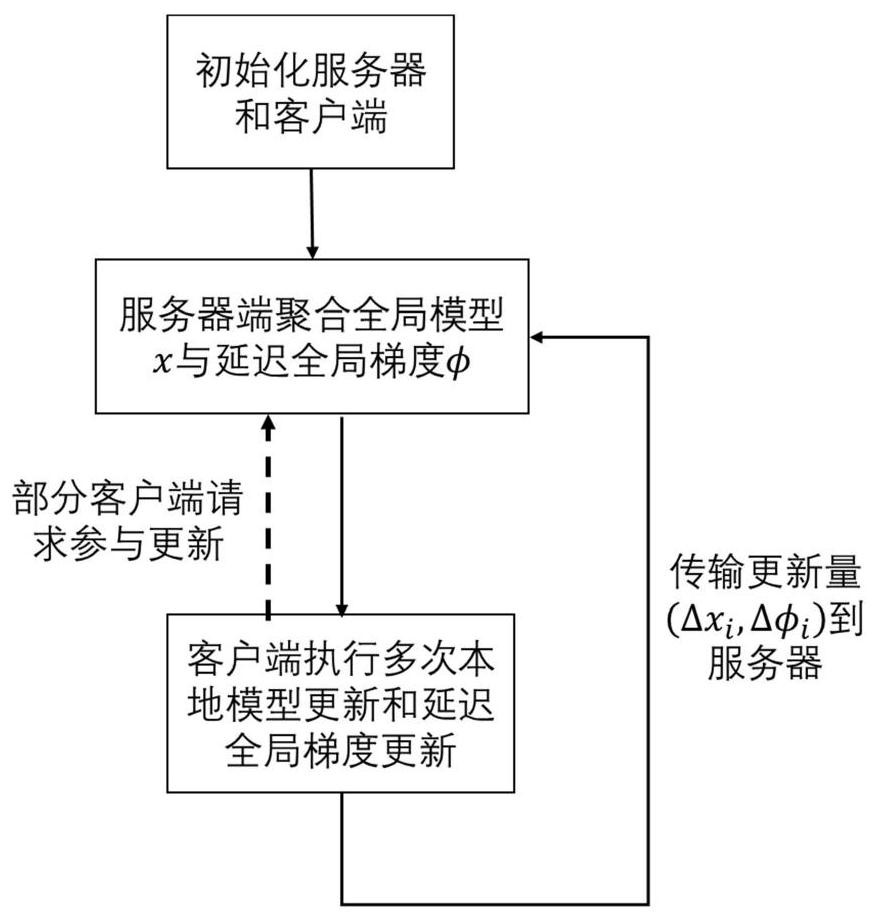
Federal learning optimization method and device
PendingCN113435604ASolve deviationReduce stochastic gradientMachine learningMachine learningFederated learning
The invention provides a federal learning optimization method and device. The method comprises the steps: obtaining a global model and a delayed global gradient which are transmitted by a server side in the current round of federated learning, wherein the delayed global gradient is obtained by updating the global gradient of the last round based on respective local data in the last round of federal learning; on the basis of the global model and the delayed global gradient of the current round, updating the local model through the local data, obtaining the federated learning update quantity, wherein the federal learning update quantity comprises the update quantity of the local model and the update quantity of the delayed global gradient; sending the federal learning update quantity to the server side. Therefore, the server side performs information aggregation according to the federated learning update quantity to obtain a new global model and a new global gradient, and sending the new global model and the new global gradient to each client side for next round of federated learning. The method effectively overcomes the problem of model deviation, improves the communication efficiency, and reduces the calculation complexity.
Owner:TSINGHUA UNIV
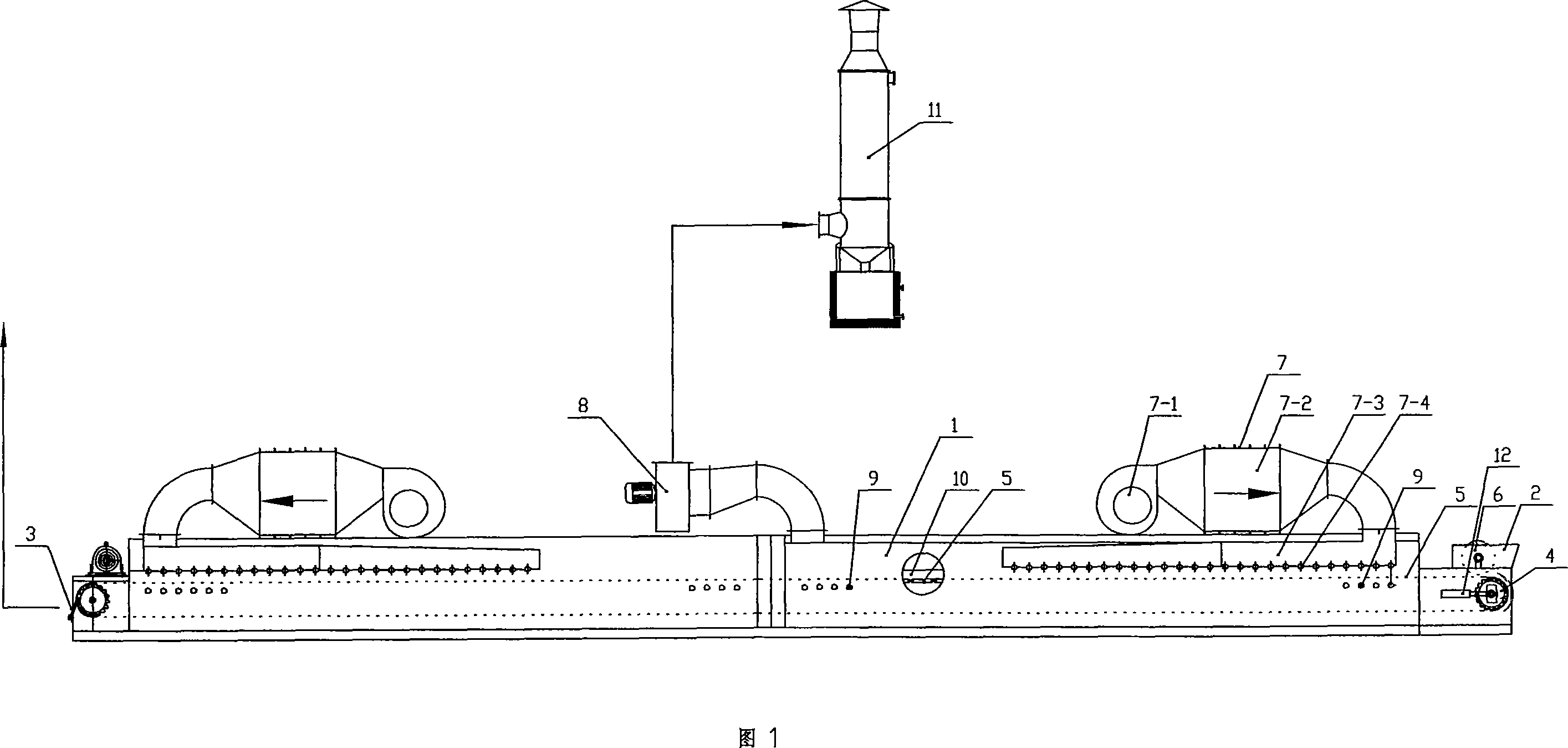
Steel belt type dryer
InactiveCN101105363ASolve deviationEasy to storeDrying machines with progressive movementsDrive wheelEngineering
The invention relates to a mechanical device, in particular to a steel belt type dryer. The steel belt type dryer has a drying passage, a feed port and a discharge port are respectively equipped at two ends of the drying passage, a conveyer belt driven by a driving wheel is equipped in the drying passage, the conveyer belt is a stainless steel belt, a baffle is respectively equipped at two sides of the conveyer belt, a feeding mechanism is equipped at the feed port, and the feeding mechanism is a feeding press roller made from rubber or silicone rubber. A gas heating device and a dehumidifying blower are connected on the drying passage. The invention can be used for drying viscous, paste-like materials with nanometer degree of fineness. The invention has the advantages of sufficient drying, and high drying efficiency.
Owner:查晓峰
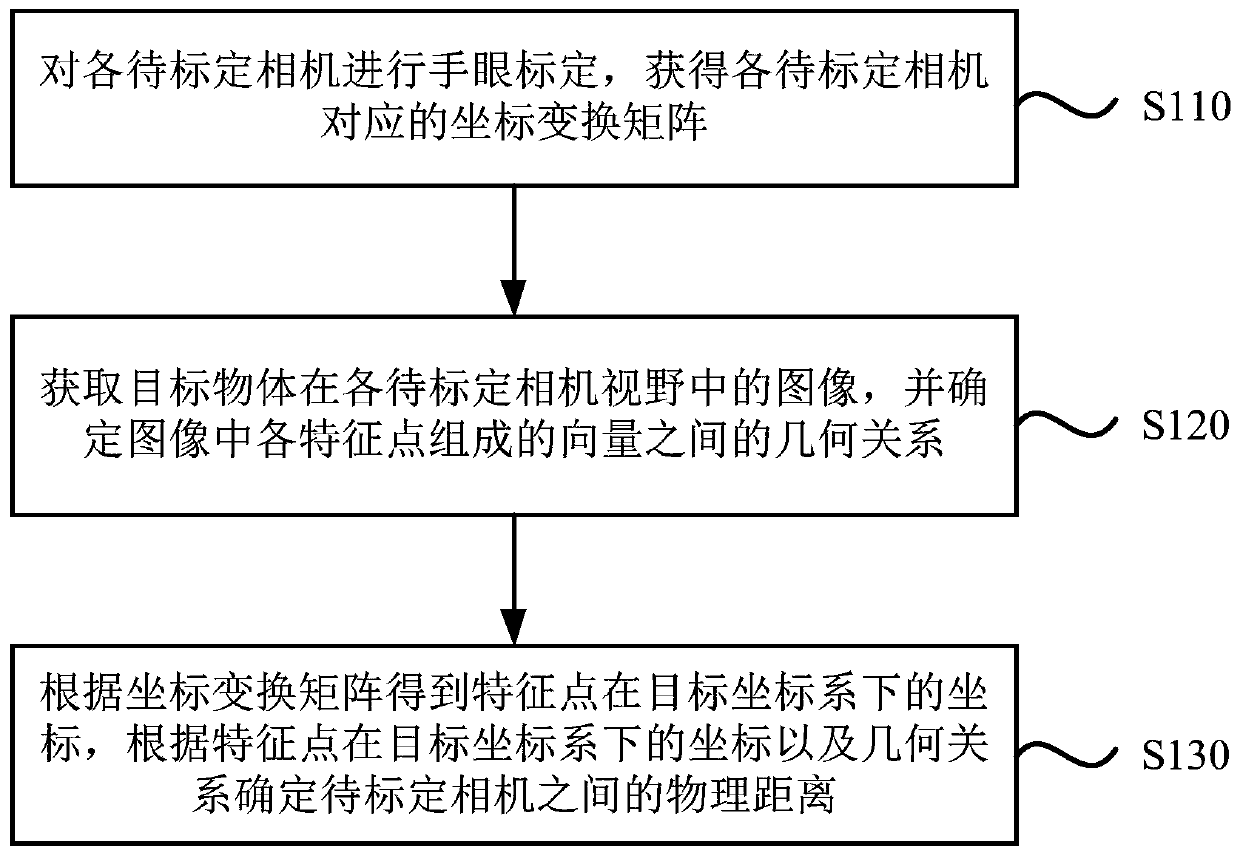
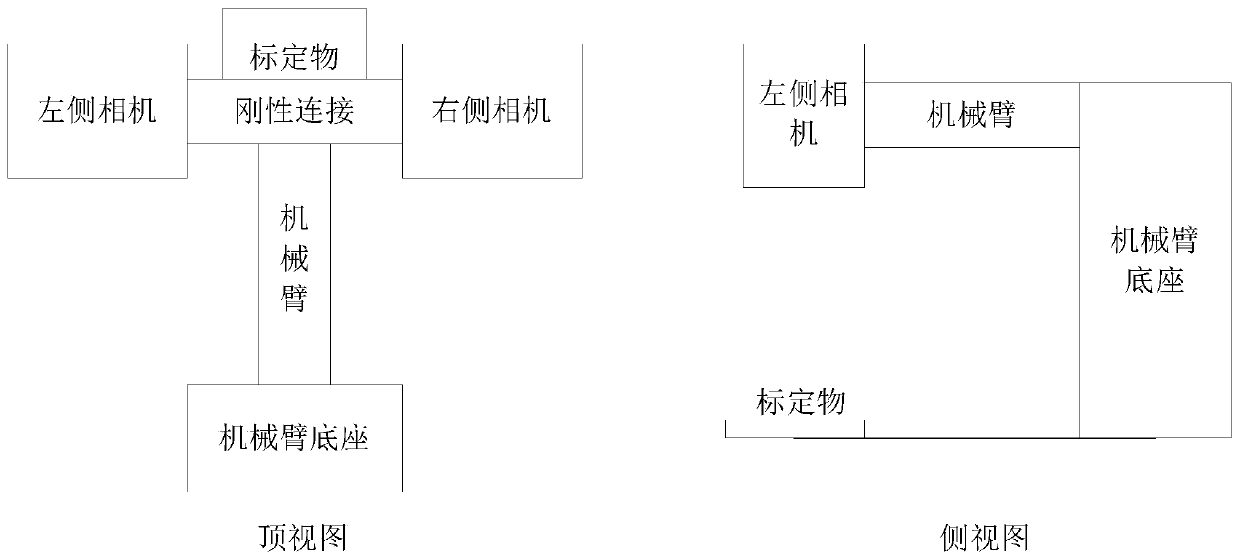
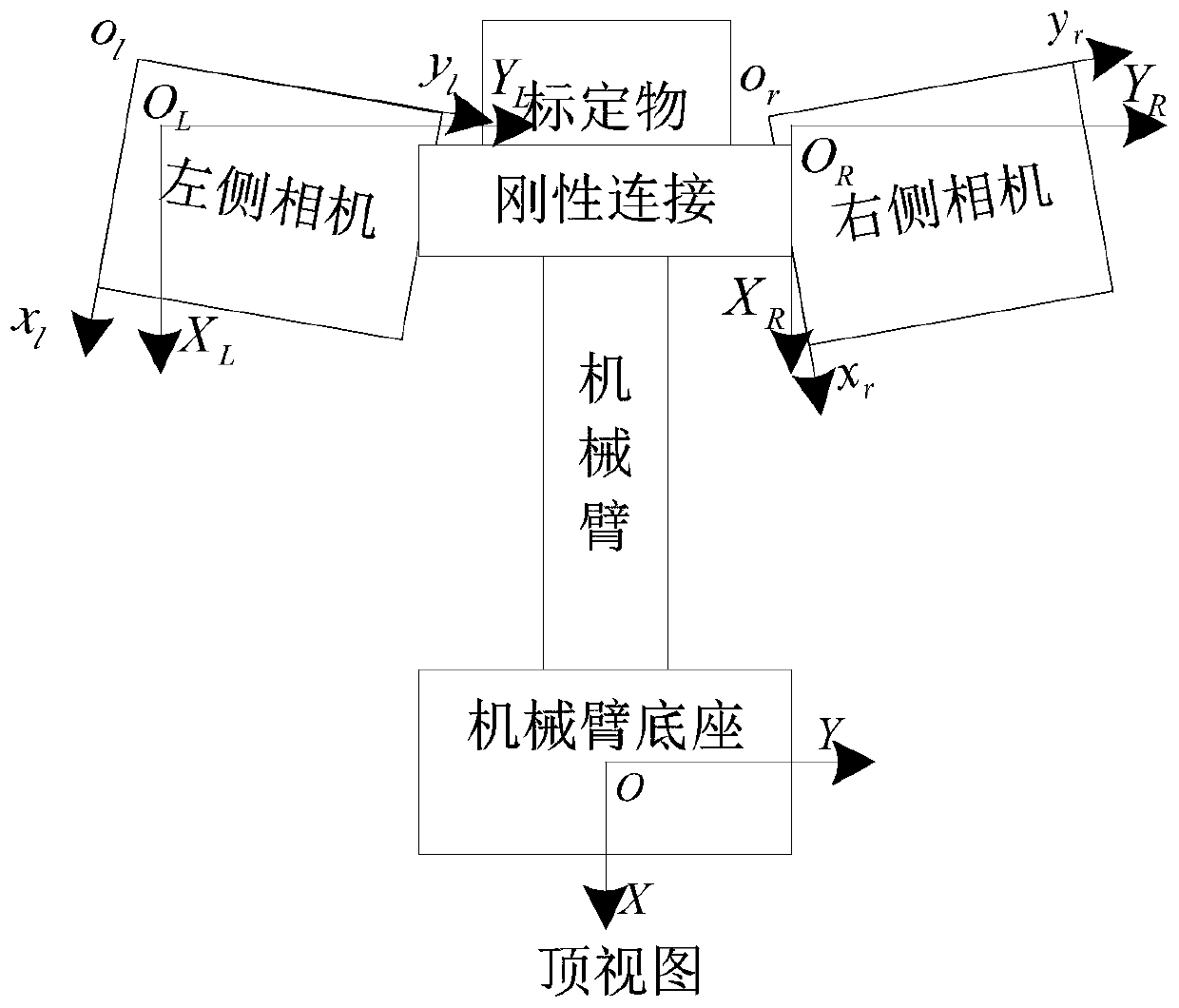
Binocular camera calibration method and device, equipment and medium
PendingCN111445533AAddress Bias and ErrorsImprove accuracyImage analysisPhysicsComputer graphics (images)
The embodiment of the invention discloses a binocular camera calibration method and device, equipment and a medium. The method comprises the steps: carrying out the hand-eye calibration of all to-be-calibrated cameras, and obtaining a coordinate transformation matrix corresponding to each to-be-calibrated camera; acquiring an image of the target object in the visual field of each to-be-calibratedcamera, and determining a geometrical relationship between vectors formed by the feature points in the image; and obtaining coordinates of the feature points in the target coordinate system accordingto the coordinate transformation matrix, and determining a physical distance between the to-be-calibrated cameras according to the coordinates of the feature points in the target coordinate system andthe geometrical relationship. According to the binocular camera calibration method provided by the embodiment of the invention, the coordinates of the feature points under the target coordinate system are obtained according to the coordinate transformation matrix corresponding to each to-be-calibrated camera; and the physical distance between the cameras to be calibrated is obtained by combiningthe geometrical relationship between the vectors formed by the feature points, so that the deviation and error caused by manual installation and measurement are solved, and the calibration accuracy ofthe binocular camera is improved.
Owner:GUANGDONG BOZHILIN ROBOT CO LTD
3D printing wire feeding, clamping and guiding device
InactiveCN105690771AImprove clamping bite forceIncrease frictionAdditive manufacturing apparatusEngineeringGuide wires
The invention discloses a 3D printing wire feeding, clamping and guiding device, and relates to the technical field of quick molding of 3D printing. The 3D printing wire feeding, clamping and guiding device comprises a guide wheel I and a guide wheel II; the guide wheel I and the guide wheel II are arranged side by side; a lap of V-shaped tooth grooves I, inwards recessed, is formed in the peripheral surface of the guide wheel I; a lap of V-shaped tooth grooves II, inwards recessed, is formed in the peripheral surface of the guide wheel II; the V-shaped tooth grooves I in the guide wheel I and the V-shaped tooth grooves II in the guide wheel II are opposite in position; when the guide wheel I and the guide wheel II are arranged side by side, the V-shaped tooth grooves I and the V-shaped tooth grooves II surround guide grooves for clamping and guiding wires; waste grooves I communicated with the V-shaped tooth grooves I are inwards recessed in the inner sides of the V-shaped tooth grooves I; and waste grooves II communicated with the V-shaped tooth grooves II are inwards recessed in the inner sides of the V-shaped tooth grooves II. The 3D printing wire feeding, clamping and guiding device can improve the wire surface clamping engagement force and friction force to prevent the wire slip or deviation in the feeding process.
Owner:GUANGXI UNIVERSITY OF TECHNOLOGY
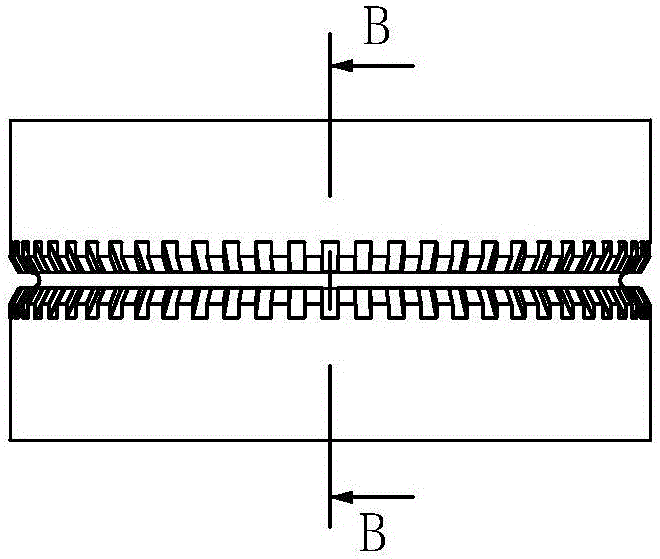
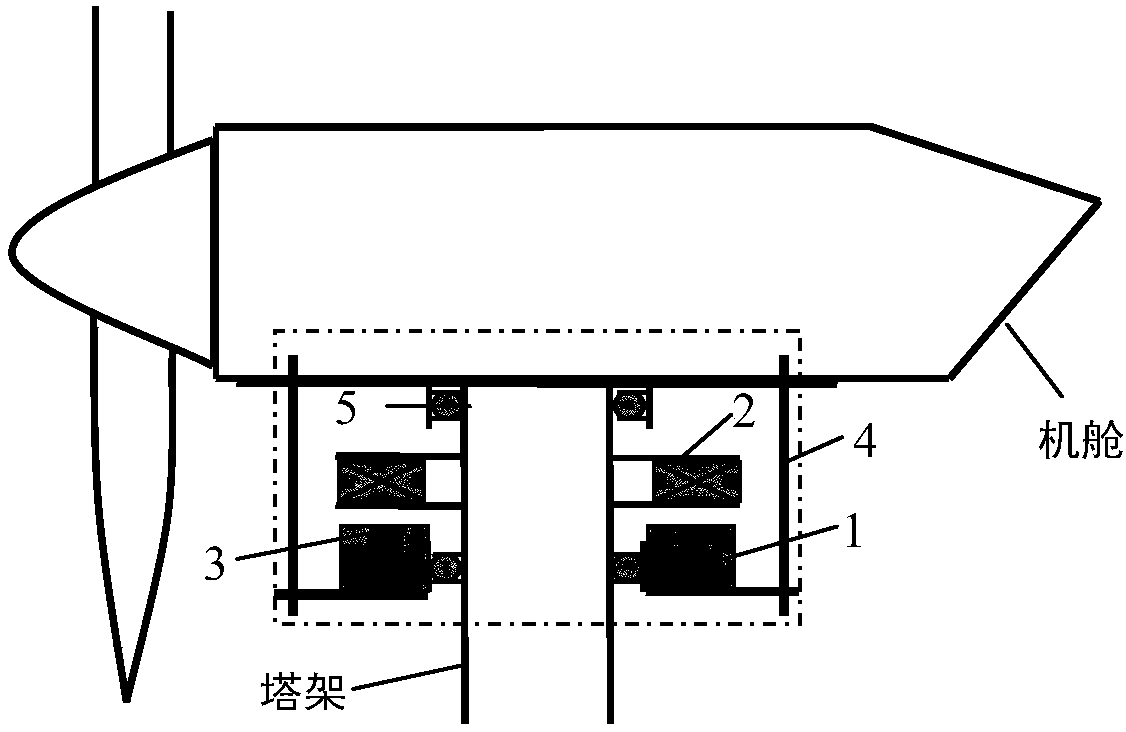
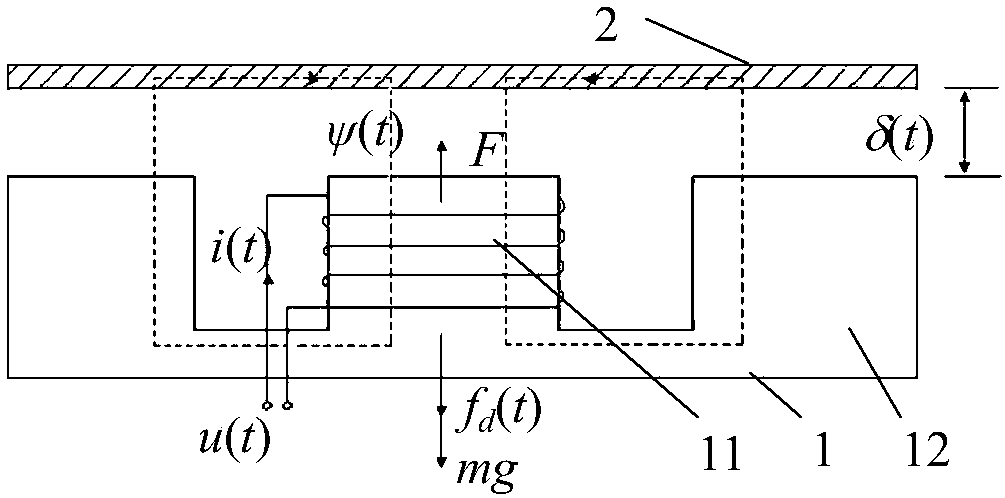
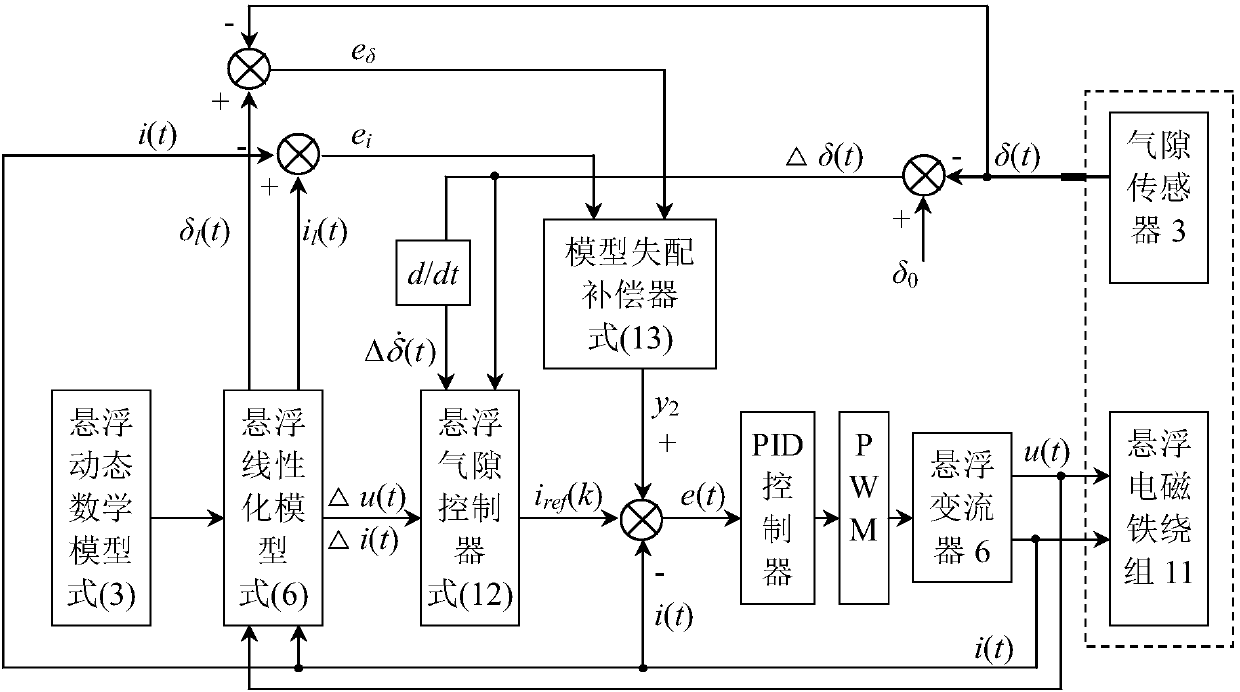
Wind power magnetic suspension yaw system suspension control method based on model mismatch compensator
ActiveCN108488036ASuspension stabilityHigh control precisionWind motor controlMachines/enginesElectrical engineering technologyYaw system
The invention relates to a wind power magnetic suspension yaw system suspension control method based on a model mismatch compensator and belongs to the technical field of electric engineering. According to the method, by means of combination of model prediction control, PID and a model mismatch device, the problem of deviation caused by model mismatch due to linearized processing close to a magnetic suspension yaw system balance point is solved, and suspension at the balance point is controlled stably in real time; a model prediction control strategy is adopted according to a linearized model,a suspension air gap controller is designed, and outer ring suspension air gap control is achieved; output of the suspension air gap controller and output of the model mismatch device are added, a current measurement value of a suspension electromagnet winding is subtracted, a PID controller controls currents of the winding, and inner ring current tracking control is achieved; and suspension airgap deviation and winding current deviation are multiplied by respective adjusting parameters and then added to form the model mismatch device. The method is high in control precision, can effectivelysuppress the influence of external disturbance, and ensure that suspension close to the balance point is fast and stable.
Owner:QUFU NORMAL UNIV
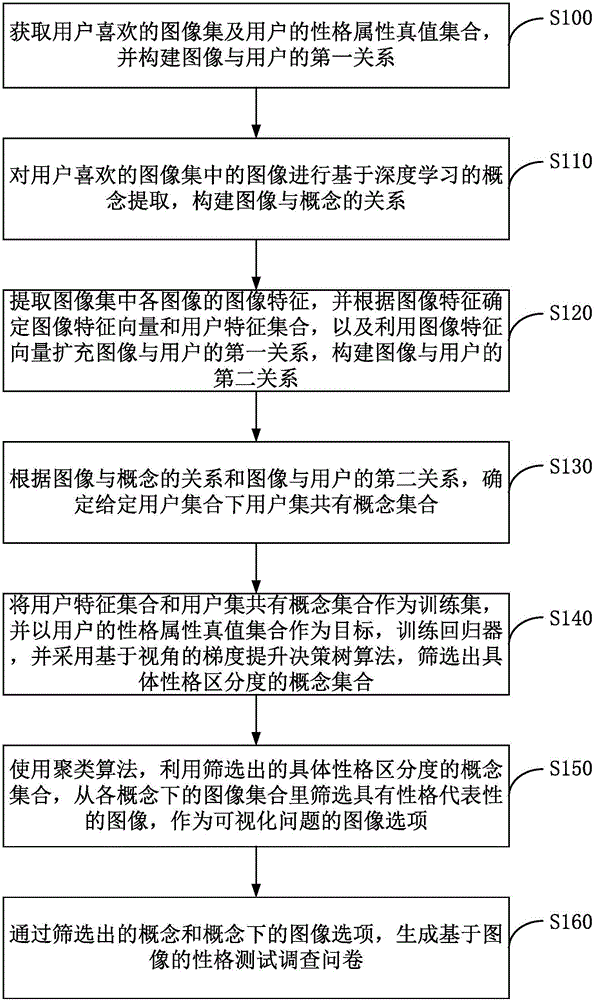
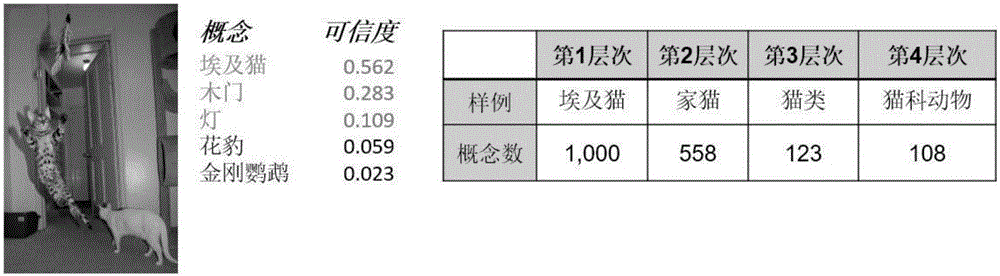
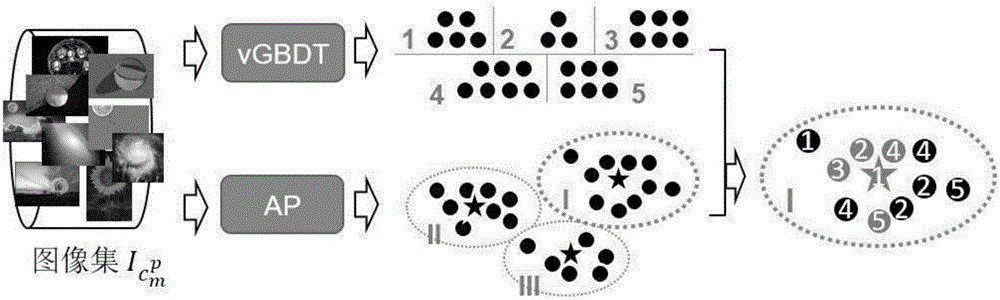
Method for generating character test questionnaire based on image and surveying interactive method
InactiveCN105893784AReduce the burden onImprove friendlinessSpecial data processing applicationsElectronic clinical trialsTruth valueImaging Feature
The invention discloses a method for generating a character test questionnaire based on an image and a surveying interactive method. The method for generating the character test questionnaire comprises the following steps: acquiring an image set which the user likes and a character nature truth set of the user and establishing a first relationship between the image and the user; extracting a concept and establishing the relationship between the image and the concept; extracting image features of each image in the image set and establishing a second relationship of the image and the user; confirming a concept set shared by a user set under a given user set according to the relationship between the image and the concept and the relationship of the image and the user; screening out the concept set at specific character distinguishing degree; and screening out the image with representative character from the image set under each concept to be taken as an image option of a visualization problem; generating the questionnaire through the screened-out concept and the image option under the concept. According to the embodiment of the invention, the accurate user character can be acquired within a shorter time period and the cross-language property is better.
Owner:INST OF AUTOMATION CHINESE ACAD OF SCI
Belt conveyor with telescopic head
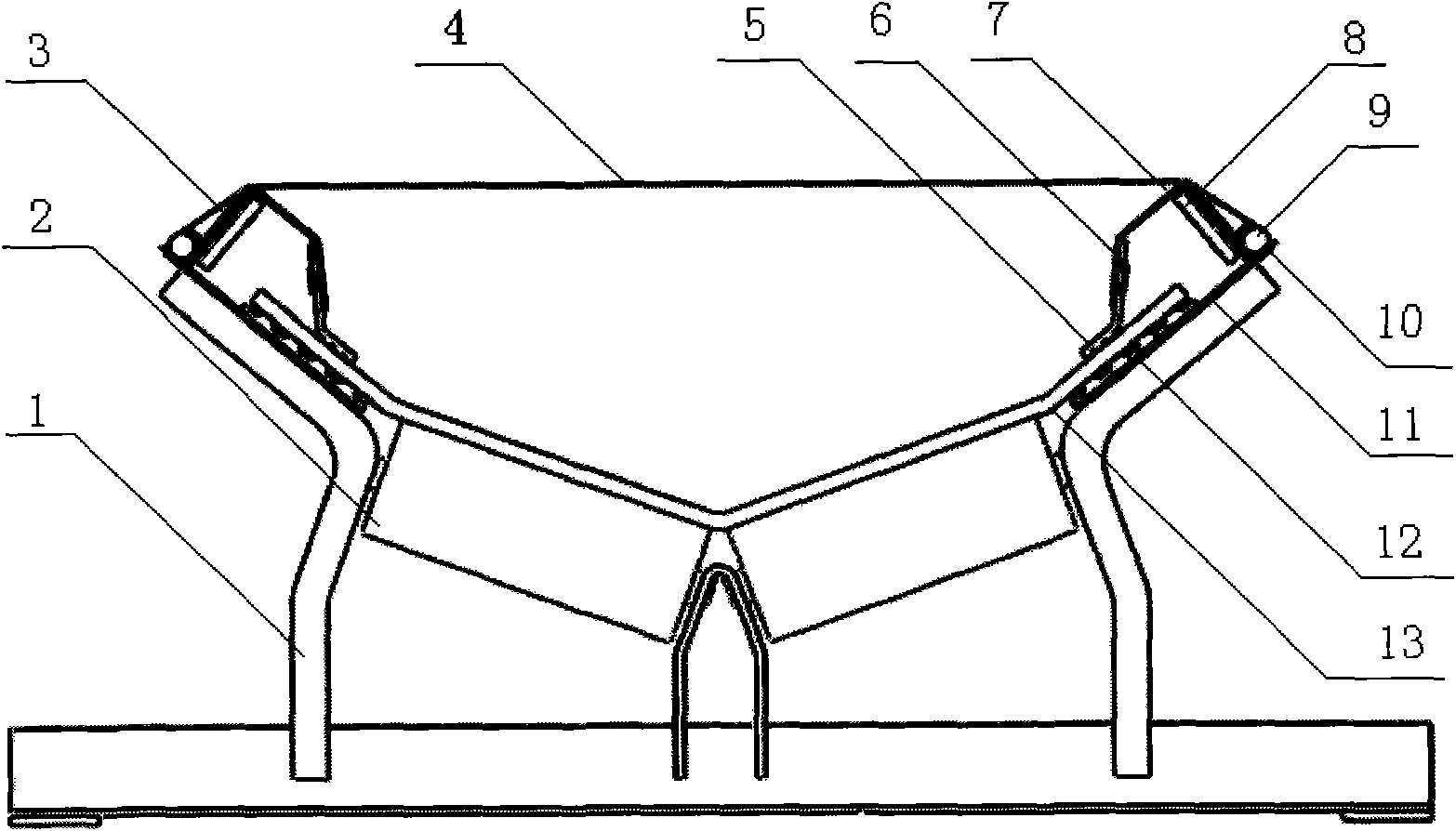
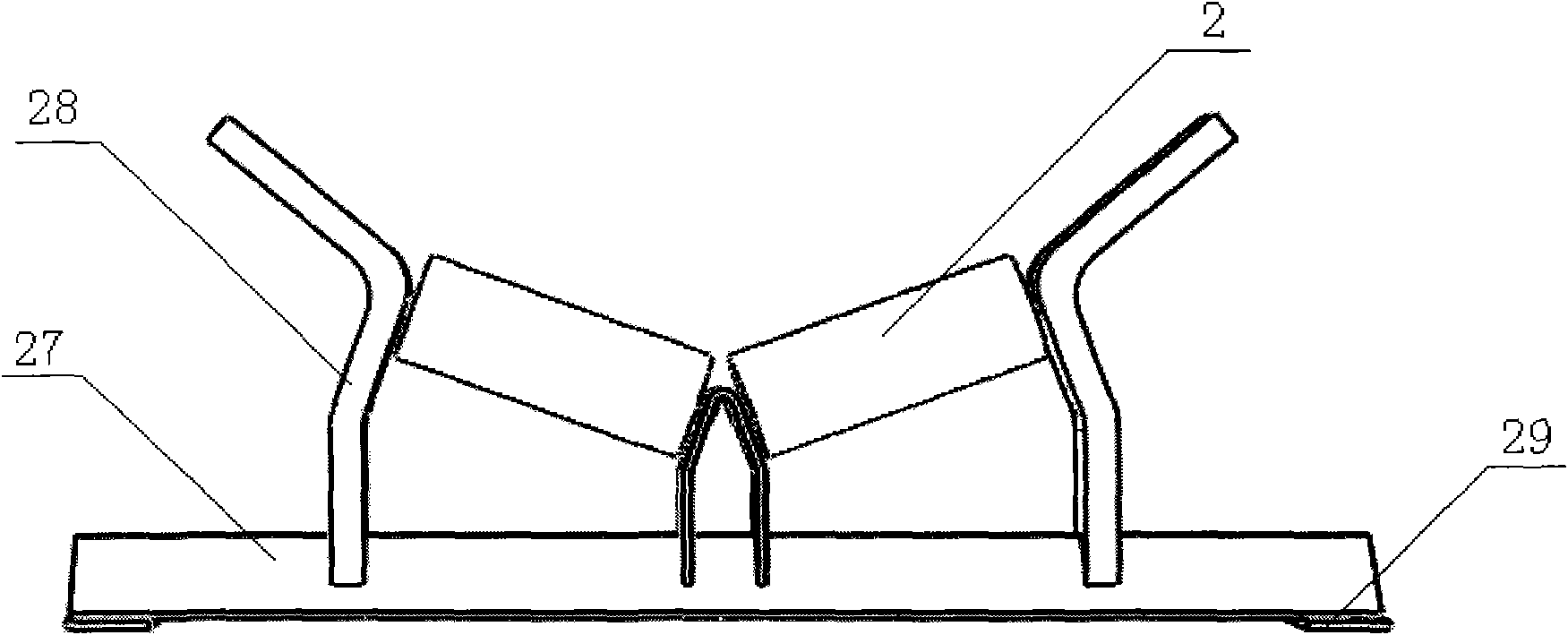
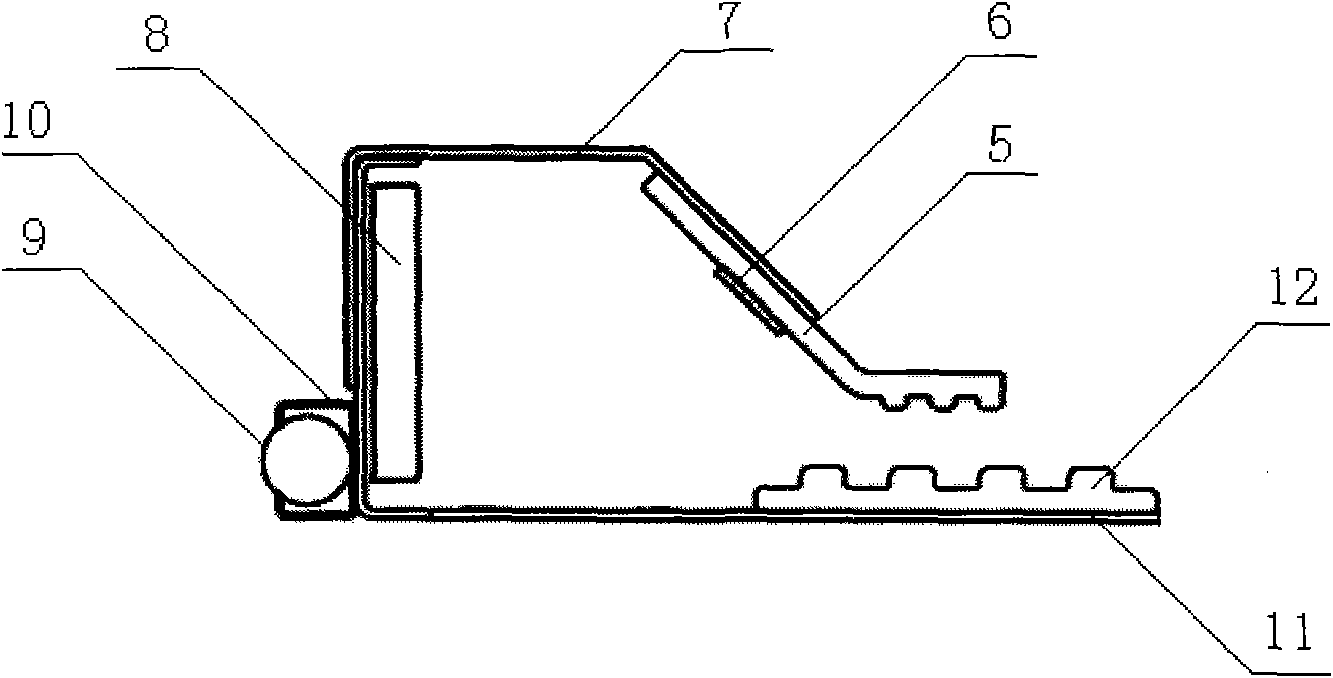
The invention discloses a belt conveyor with a telescopic head. The belt conveyor comprises a frame with one end arranged into a truss body, a walking mechanism arranged at the bottom of the frame, a guide mechanism arranged on the truss body and the frame and a gear and rack transmission device driving the frame to move in the truss body. A bearing support roll is fixed at the upper end of the frame, a head roller is fixed on the head portion of the frame, a first redirection roller is fixed on the tail portion of the frame, and a second redirection roller is fixed at the bottom of the truss body. Due to the fact that the head of the belt conveyor can be retreated, collision between the head and a moving device can be avoided, and the operation efficiency of the moving device is improved. In addition, the blanking central position of the head is adjustable, and the off tracking problem of a sealing-tape machine is solved.
Owner:TONGLING MIRACLE MECHANICAL EQUIP
Sunshading curtain for automobile
Owner:陈海龙
Self-moving belt conveyor tail
ActiveCN103410561AReduce the number of demolitionImprove work efficiencyEarth drilling and miningUnderground transportDislocationBelt conveyor
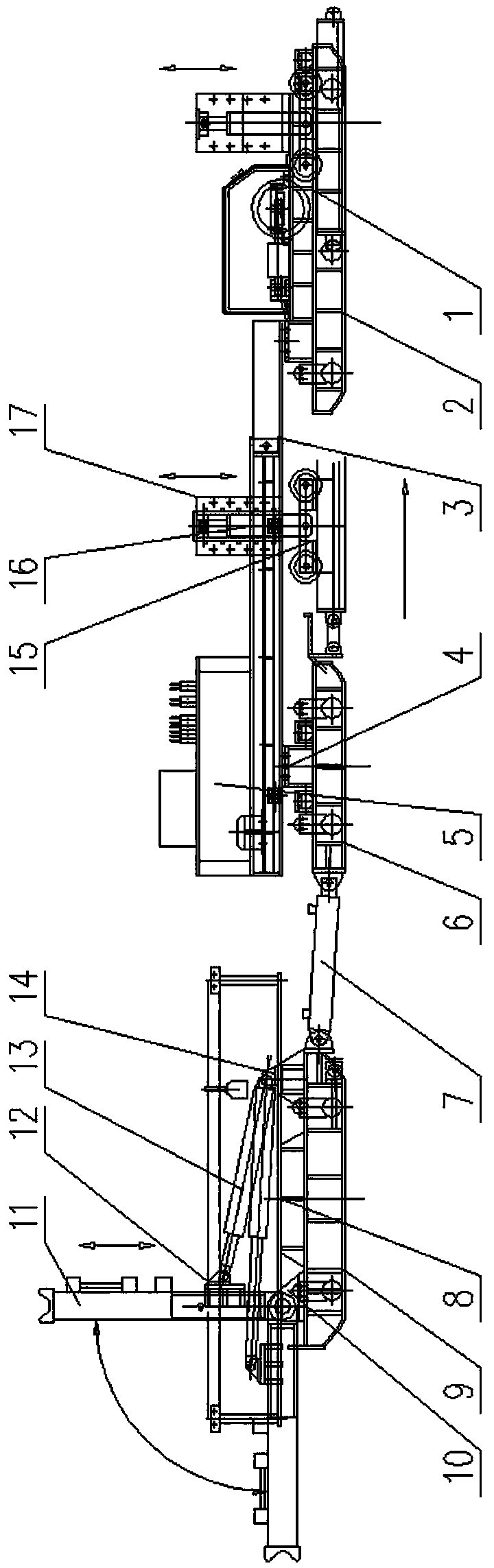
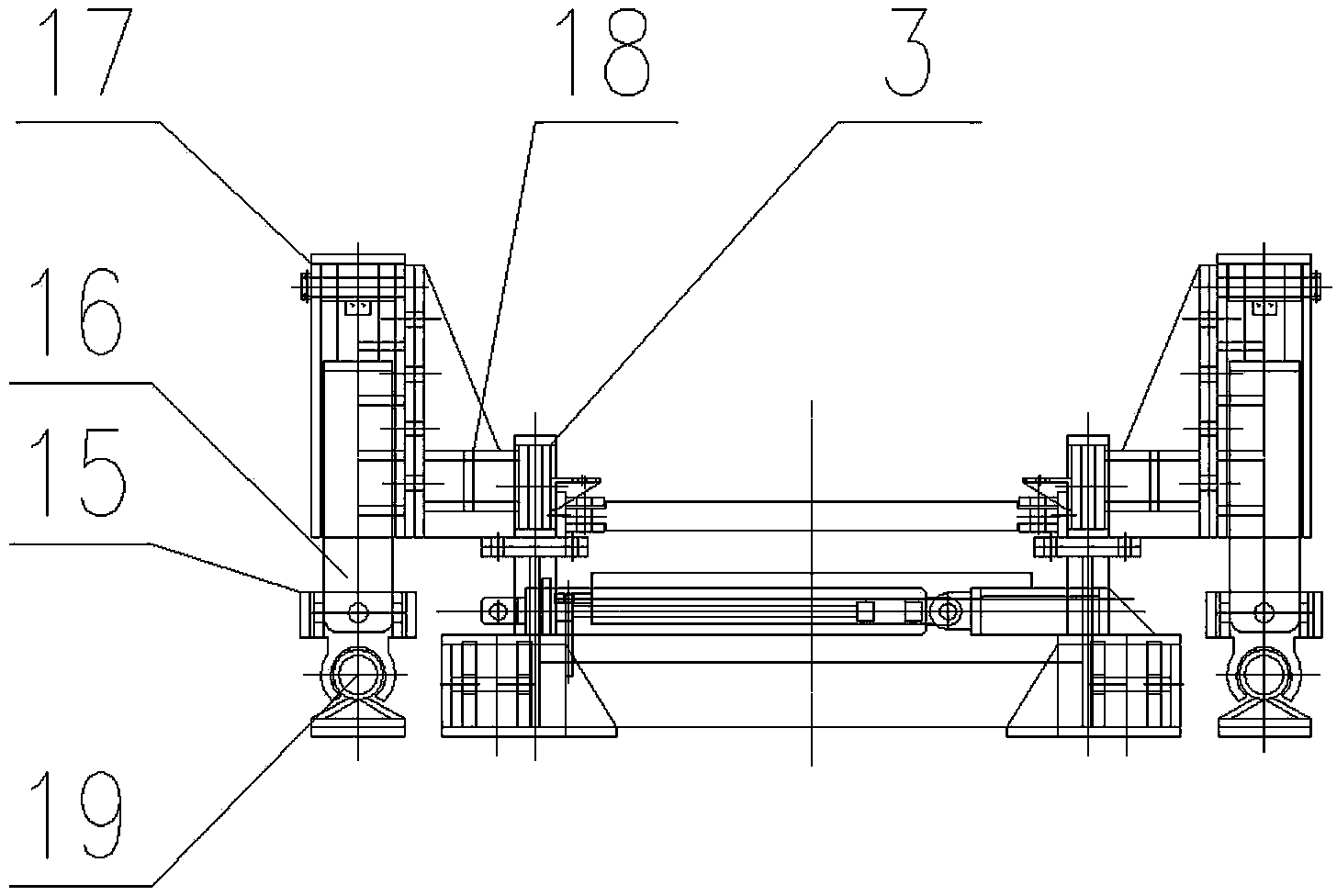
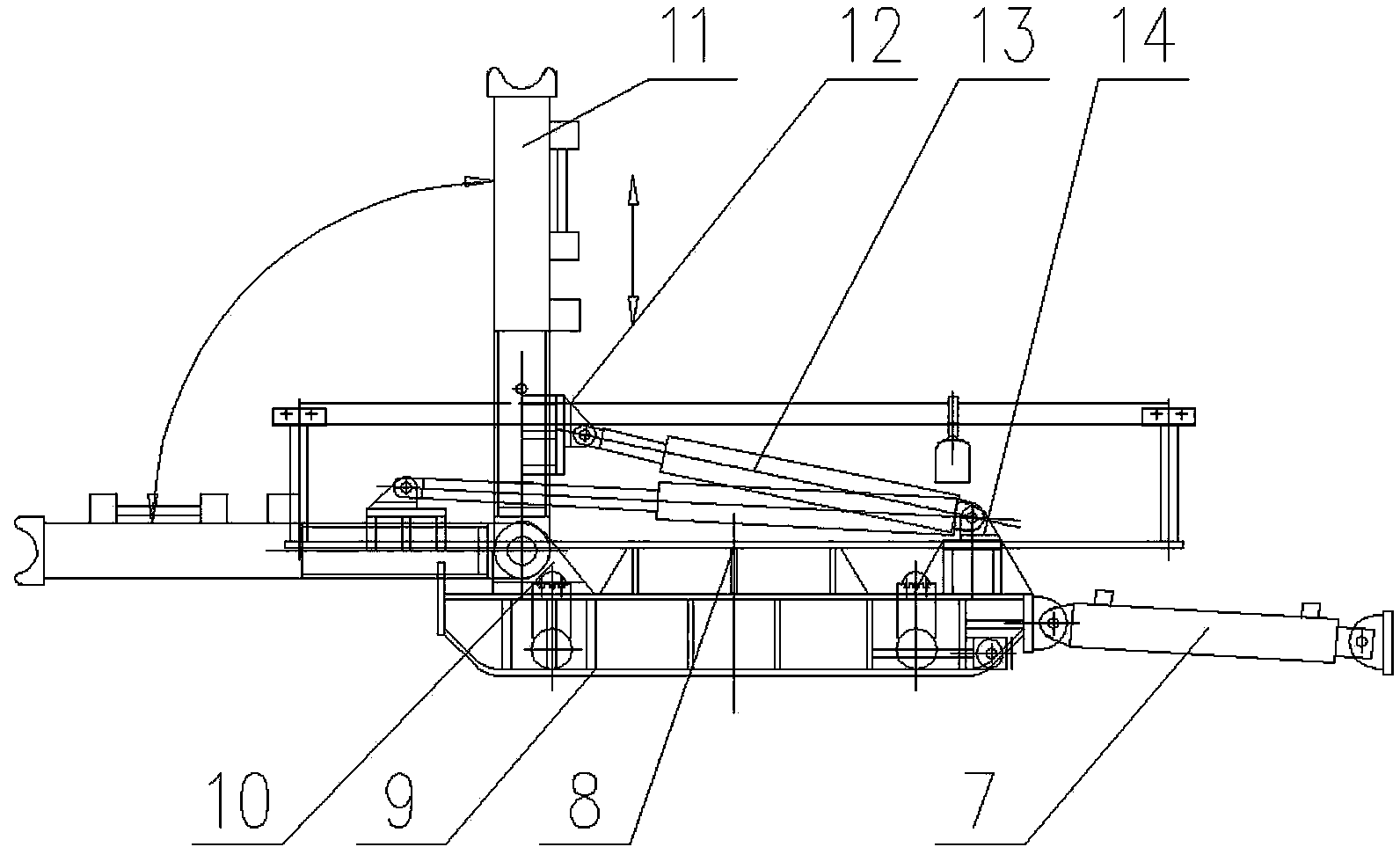
The invention relates to belt conveyor moving devices, particularly to a self-moving belt conveyor tail. The self-moving belt conveyor tail comprises a conveyor tail, a middle portion, a driving portion and a hydraulic system and is characterized by further comprising a moving oil cylinder, one end of the moving oil cylinder is hinged to the base of the driving portion, the other end of the moving oil cylinder is hinged to the middle portion, and a beam arranged on a middle portion base is connected with a conveyor tail base. The self-moving belt conveyor tail further comprises tracks which are arranged on two sides of the conveyor tail and the middle portion, and the rear ends of the tracks are connected with the base of the driving portion; the middle portion and the conveyor tail portion is provided with a lifting mechanism, the driving portion is provided with an anti-slipping fixing mechanism which can personally achieve fixation to prevent dislocation of the conveyor tail during transportation, the middle portion is provided with an offset regulating mechanism which can solve the off-tracking problem of belts, the conveyor tail is provided with a roller offset regulating mechanism which can solve the deviation of a conveyor tail roller. The self-moving belt conveyor tail can move by itself, each movement distance maximally reaches 1.8 meters, the lifting mechanism and the moving oil cylinder enable the conveyor tail and the middle portion to move along the tracks; and the tacks can smoothly move in two side tunneling directions of the conveyor tail portion.
Owner:山西潞安环保能源开发股份有限公司常村煤矿
Digital finder method employing geostationary satellite of stationary orbit
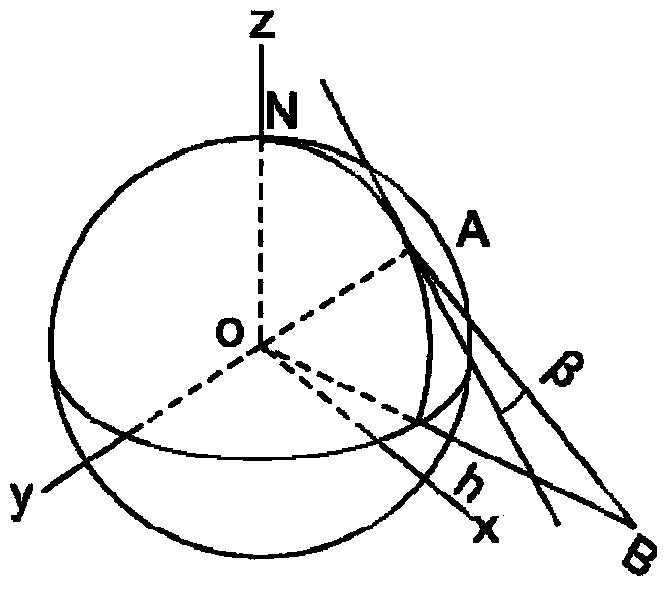
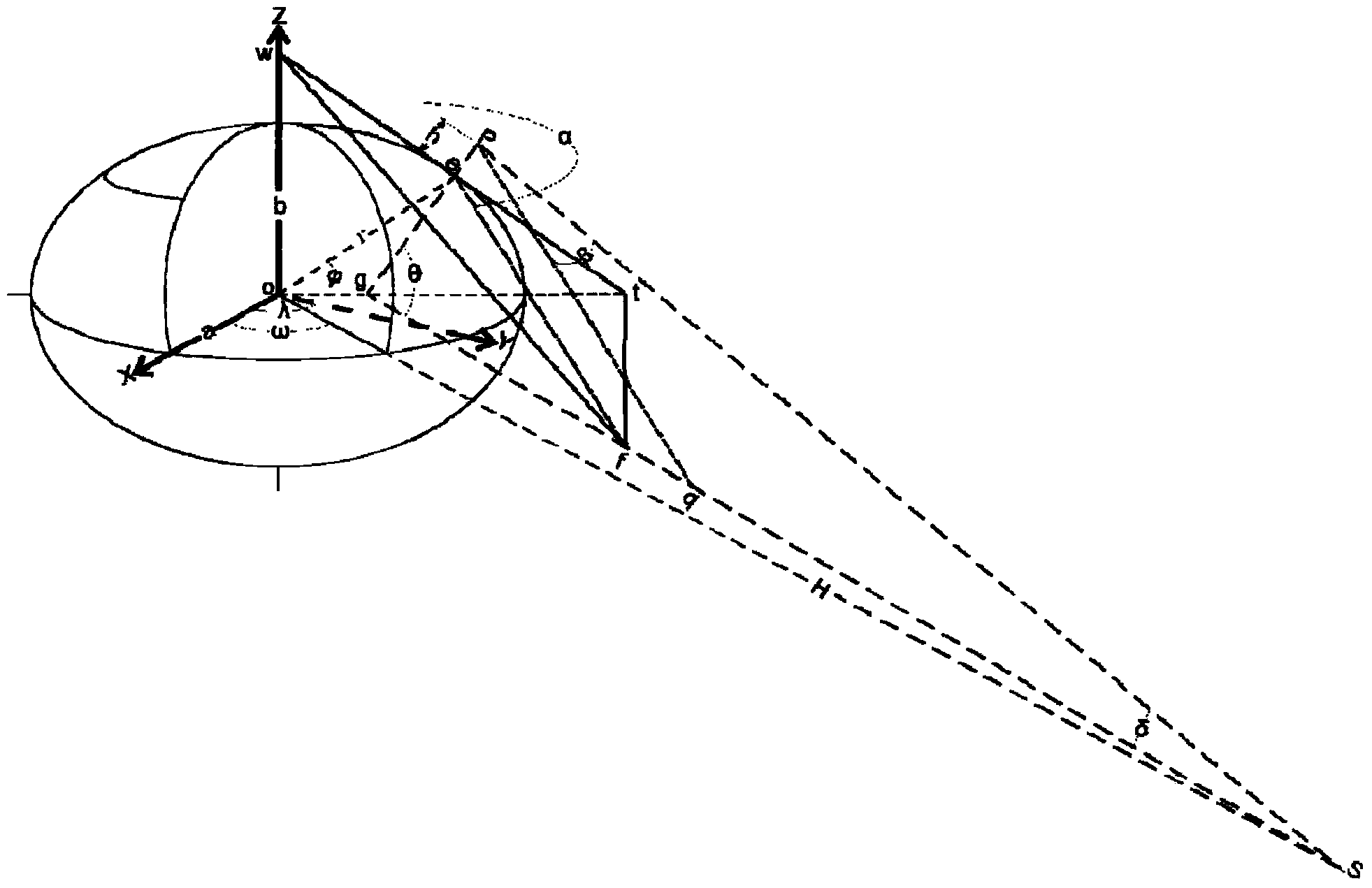
InactiveCN103727919ASolve deviationAvoid blindnessAngle measurementOptical rangefindersGreek letter betaInteraction interface
The invention relates to the technical field of a geostationary satellite of a stationary orbit, and particularly discloses a digital finder method employing a geostationary satellite of a stationary orbit. The digital finder method comprises the following steps: directly collecting position information of a ground point through a global position system (GPS) or Beidou terminal equipment; inputting parameters of position information of the ground point through a call instruction based on a http protocol; calculating through a position processing module of the appointed ground point, an ellipsoid secondary variable calculation module, a sighting distance processing module from the appointed ground point to the appointed satellite, an elevation processing module from the appointed ground point to the appointed satellite, and a true north azimuth processing module from the appointed ground point to the appointed satellite; solving the results of a sighting distance L from any ground point to the satellite, the elevation beta from any ground point to the satellite and an azimuth angle alpha from any ground point to the satellite; carrying out conversion through a data conversion module, displaying the finder result in a human-computer interaction interface, and determining the relative positions of the ground point and the satellite through the finder result. The digital finder method is rigorous in theory, efficient, and high in calculating result accuracy.
Owner:XIAN CENT OF GEOLOGICAL SURVEY CGS +1
Integrated storage cabinet with flexible material distribution effect
InactiveCN102530578AMeet proportional mixing needsNot easy to bend and deformTobacco preparationLoading/unloadingReciprocating motionEngineering
The invention discloses an integrated storage cabinet with the flexible material distribution effect, which comprises a storage cabinet (5), a longitudinal reciprocating trolley (2), a transversal reciprocating trolley (1) and a conveyer belt, wherein L-shaped chain tracks (18) are symmetrically arranged on the inner side of a storage cabinet frame (3) and extend in the longitudinal direction of the storage cabinet, a conveyor chain belt (19) is arranged on the chain tracks, the two sides of a supporting bar (8) which is positioned at the bottom of the storage cabinet are arranged on a conveyer chain (11) of the conveyor chain belt, and a chain driving device (16) drives the conveyer chain to move the conveyor chain belt on the chain tracks; the width of the storage cabinet is 1400 mm to 5500 mm, and the storage capacity is 2 tons to 60 tons; a longitudinal guide track (6) is arranged at the top of the storage cabinet, and the longitudinal reciprocating trolley is positioned on the longitudinal guide track and makes a reciprocating motion; a transversal guide track (4) is arranged on the frame of the longitudinal reciprocating trolley, and the transversal reciprocating trolley is positioned on the transversal guide track and driven by a multipoint driving device (21) to make a reciprocating motion and selectively distribute materials in any region in the width direction of the storage cabinet; and a support roller guide track (10) is arranged at the bottom of the storage cabinet frame, a support roller component (9) is arranged at the bottom of the supporting bar and in the position corresponding to the support roller guide track, and the support roller component is assembled on the support roller guide track. The storage cabinet can distribute materials in any position in the width direction of the storage cabinet and ensure uniform distribution of materials, meets the requirements for large-formula proportional mixing of materials, has simple structure and low cost, and runs reliably.
Owner:YUNNAN KUNMING SHIPBUILDING DESIGN & RESEARCH INSTITUTE
Horizontal sorting machine
The invention discloses a horizontal sorting machine. The horizontal sorting machine comprises a belt conveyor, a swing arm frame arranged on the side of the belt conveyor, a swing arm fixed base, a box body and a servo motor, a servo motor reducer and a transmission mechanism arranged in the box body, wherein the servo motor respectively controls the belt conveyor or the swing arm frame to act to finish sorting operation through the opening and closing of an electromagnetic clutch in the transmission mechanism and the connection and separation of a connecting piece and a fourth shaft. According to the invention, the servo motor is adopted for driving, the frequency response is rapid, the speed is stable, the angle and speed of a swing arm are exact, and the horizontal sorting machine is energy-saving; the rotation of the swing arm and the movement of the belt conveyor are driven by a same power source, the structure is compact, the size is small, the cost is low, and the fine design is convenient; the belt conveyor adopts a roller mechanism with a groove, so that a problem that the belt conveyor is deviated because of the gravity of a belt is prevented; the transmission mechanism is arranged in the box body, so that the horizontal sorting machine is safe and clean.
Owner:朗奥(启东)自动化设备有限公司
Compact type belt conveyor
The invention discloses a compact type belt conveyor. The compact type belt conveyor comprises a first H-shaped rack, a second H-shaped rack, a drive roller and a reversing roller. Sliding rails are arranged on the inner side face of the second H-shaped rack. The first H-shaped rack can slide in the sliding rails. The drive roller is arranged at one end of the second H-shaped rack, and the reversing roller is arranged at one end of the first H-shaped rack. The compact type belt conveyor further comprises a screw nut mechanism. The screw nut mechanism comprises a screw and a nut block rotationally arranged on the screw in a sleeving manner. The second H-shaped rack is provided with a penetrating hole matched with the screw in diameter. The second H-shaped rack is further fixedly provided with a plate with a limiting groove, the nut block is located in the limiting groove, and the nut block is further provided with a protruding part which stretches out of the limiting groove and is integrally formed with the nut block. A rod is hinged to the protruding part, and the other end of the rod is hinged to the first H-shaped rack. The compact type belt conveyor is simple and compact in structure, capable of achieving tensioning or deviation correcting, and suitable for being used on occasions with the small space.
Owner:贵州通顺矿山机械有限公司
High-efficiency ground sampling device and sampling method
ActiveCN109085014AQuickly meet the demand for sampling volumeReduce time consumptionWithdrawing sample devicesDrive wheelDrive shaft
The invention discloses a high-efficiency ground sampling device and a sampling method, and belongs to the field of geotechnical exploration equipment. The device and the method have an advantage thatthe sampling efficiency is improved. According to the technical scheme, the high-efficiency ground sampling device comprises a fixed support component, a gasoline engine, a transmission component anddrilling components, wherein the gasoline engine, the transmission component and the drilling components are arranged on the fixed support component and are connected in sequence; the drilling components are externally connected to a water cooling system; the gasoline engine is connected to the transmission component; the transmission component is arranged in a mounting box; a driving wheel, a driving shaft which is coaxial to the driving wheel, a driven wheel, a driven shaft which is coaxial to the driven wheel, and a transmission chain which is arranged between the driving wheel and the driven wheel are arranged in the mounting box; the driving shaft is connected to a piston of the gasoline engine through a coupling; one end, far away from the gasoline engine, of the driving shaft and one end, far away from the gasoline engine, of the driven shaft are respectively connected to a set of drilling components; and a sliding component is connected between the gasoline engine and the fixed support component, and the gasoline engine moves up and down along the fixed support component.
Owner:江苏中煤地质工程研究院有限公司
An improved unmanned formula car brake mechanism
PendingCN110228451ALower the altitudeAvoid track deviationFoot actuated initiationsAutomatic initiationsVehicle frameElectric machinery
The invention discloses an improved unmanned formula car brake mechanism comprising a bottom plate, sliding rails installed on the bottom plate, a push rod, a brake plate, a hinged support plate, a pin, a connecting rod, a rocker, a motor, a controller, a pressure sensor, a brake pedal, a brake pedal arm, pulleys, a first return spring for rail reset and a second return spring for brake plate reset. The mechanism of the invention is advantaged in that: 1, the design adopts a rigid rod with large bearing capacity, small deflection and high reliability to replace a steel wire rope; 2, a motor main shaft and a rocker are connected by a spline shaft, and the forward rotation of the motor directly drives the rocker to rotate, thereby driving the connecting rod to perform translational movementso as to drive the brake plate arm to rotate for a corresponding angle, which compensates for the defects of unsatisfactory response and inaccurate trajectories in conventional winders; and 3, four pulleys are added to the bottom of the slide rails to cooperate with two sliding grooves in the frame to enable movement, thus solving the problem of mechanism offset due that mechanisms are not compact.
Owner:GUANGDONG UNIV OF TECH
Features
- R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
Why Patsnap Eureka
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Social media
Patsnap Eureka Blog
Learn More Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com