A kind of nucleic acid aptamer of glycosylated hemoglobin and its preparation method
A technology of glycosylated hemoglobin and nucleic acid aptamer, applied in the field of protein detection, can solve the problems such as the screening preparation method has not been reported yet, the lack of research on the efficient and specific identification of glycosylated hemoglobin GHb, etc. Effects of Specificity and Sensitivity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
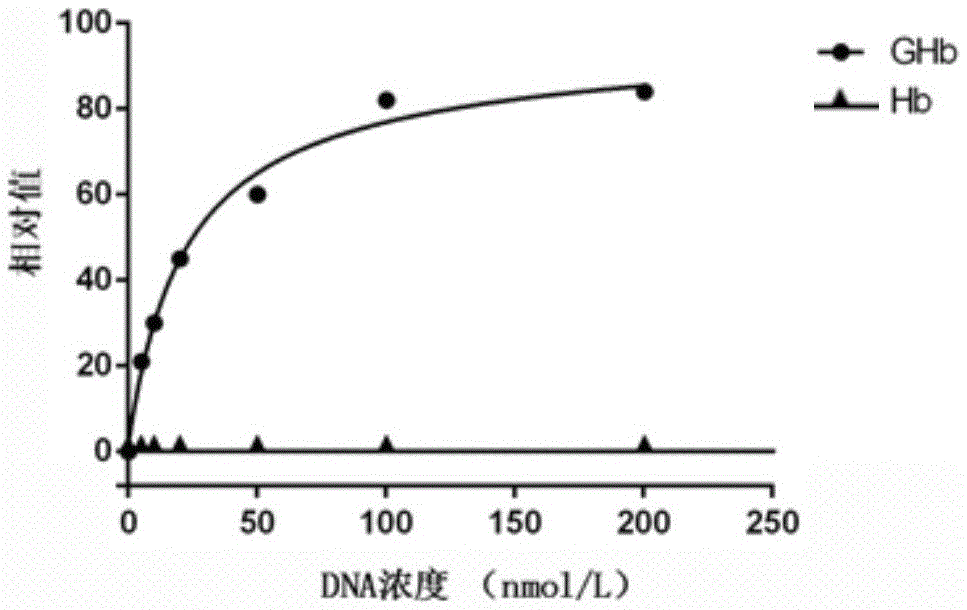
[0039] Example 1 Preparation of nucleic acid aptamer GHb1 specifically bound to glycosylated hemoglobin
[0040] (1) Construction of random sequence oligonucleotide library: artificially synthesized single-stranded DNA sequence, and the construction capacity of the library is 1×10 5 The single-stranded DNA random sequence oligonucleotide library, the DNA sequence included in the single-stranded DNA random oligonucleotide library is:
[0041] 5’-GACCATACCAGCTTATTCAATT-N 25~40 -AGATAGTAAGTGCAATCTGGC-3'
[0042] The DNA sequence of the single-stranded DNA random oligonucleotide library includes an intermediate random sequence N 25~60 and fixed sequences at both ends, the middle random sequence N 25~40 It is a random sequence of 25 to 40 bases, and the fixed sequences at both ends are: 5'-GACCATACCAGCTTATTCAATT, 3'-CGGTCTAACGTGAATGATAGA, and the fixed sequences at both ends are PCR amplification primer binding regions;
[0043] (2) Preparation of glycosylated hemoglobin-micro-...
Embodiment 2
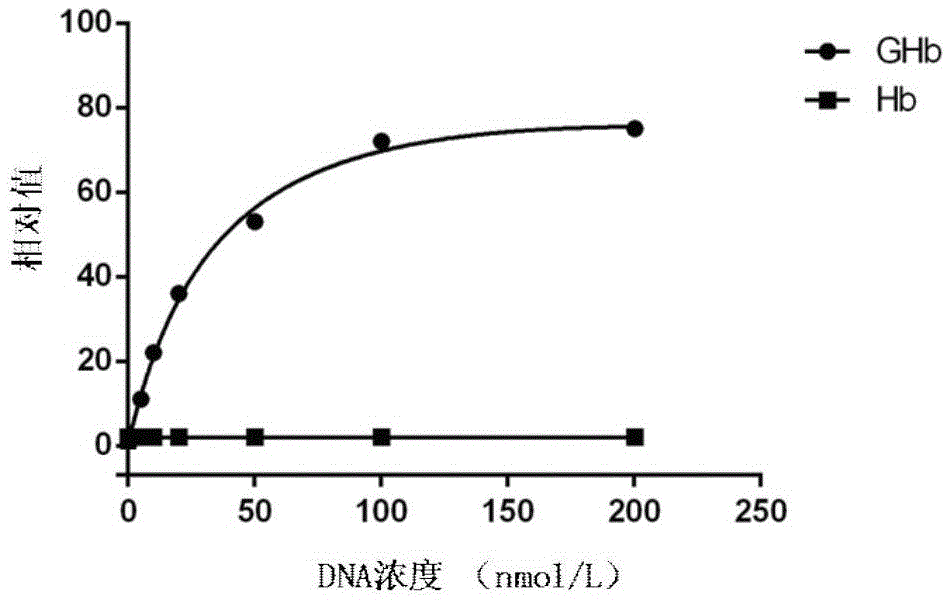
[0049] Example 2 Preparation of the nucleic acid aptamer GHb2 specifically binding to glycosylated hemoglobin
[0050] (1) Construction of random sequence oligonucleotide library: artificially synthesized single-stranded DNA sequence, and the construction capacity of the library is 3×10 5 The single-stranded DNA random sequence oligonucleotide library, the DNA sequence included in the single-stranded DNA random oligonucleotide library is:
[0051] 5’-GACCATACCAGCTTATTCAATT-N 30~45 -AGATAGTAAGTGCAATCTGGC-3'
[0052] The DNA sequence of the single-stranded DNA random oligonucleotide library includes an intermediate random sequence N 25~60 and fixed sequences at both ends, the middle random sequence N 30~45 It is a random sequence of 30 to 45 bases, and the fixed sequences at both ends are: 5'-GACCATACCAGCTTATTCAATT, 3'-CGGTCTAACGTGAATGATAGA, and the fixed sequences at both ends are PCR amplification primer binding regions;
[0053] (2) Preparation of glycosylated hemoglobin-...
Embodiment 3
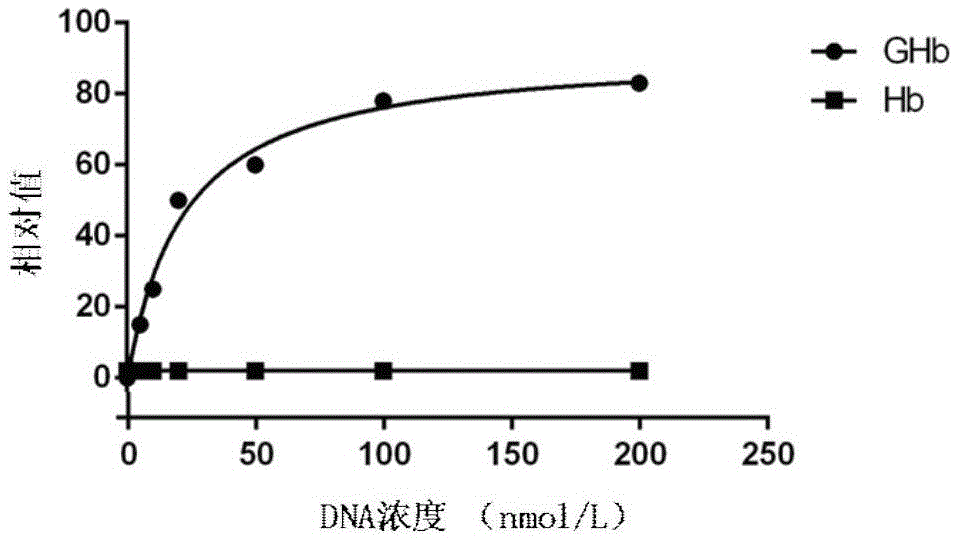
[0059] Example 3 Preparation of the nucleic acid aptamer GHb3 specifically bound to glycosylated hemoglobin
[0060] (1) Construction of random sequence oligonucleotide library: artificially synthesized single-stranded DNA sequence, and the construction capacity of the library is 1×10 6 The single-stranded DNA random sequence oligonucleotide library, the DNA sequence included in the single-stranded DNA random oligonucleotide library is:
[0061] 5’-GACCATACCAGCTTATTCAATT-N 35~60 -AGATAGTAAGTGCAATCTGGC-3'
[0062] The DNA sequence of the single-stranded DNA random oligonucleotide library includes an intermediate random sequence N 25~60 and fixed sequences at both ends, the middle random sequence N 35~60 It is a random sequence of 35 to 60 bases, the fixed sequences at both ends are: 5'-GACCATACCAGCTTATTCAATT, 3'-CGGTCTAACGTGAATGATAGA, and the fixed sequences at both ends are PCR amplification primer binding regions;
[0063] (2) Preparation of glycosylated hemoglobin-microm...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com