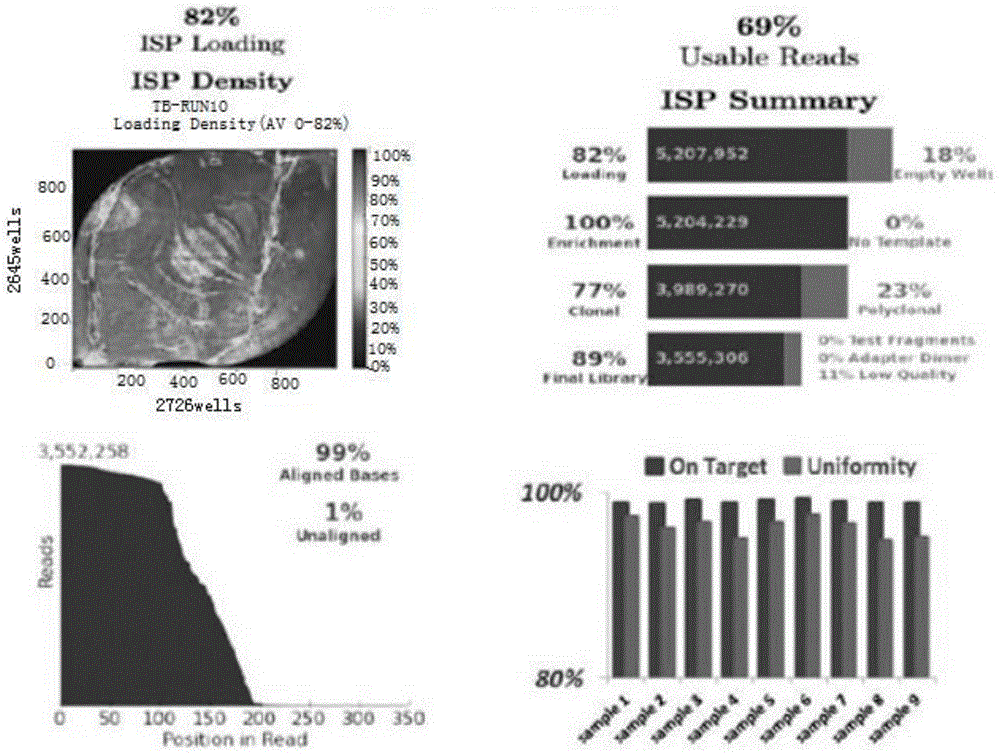
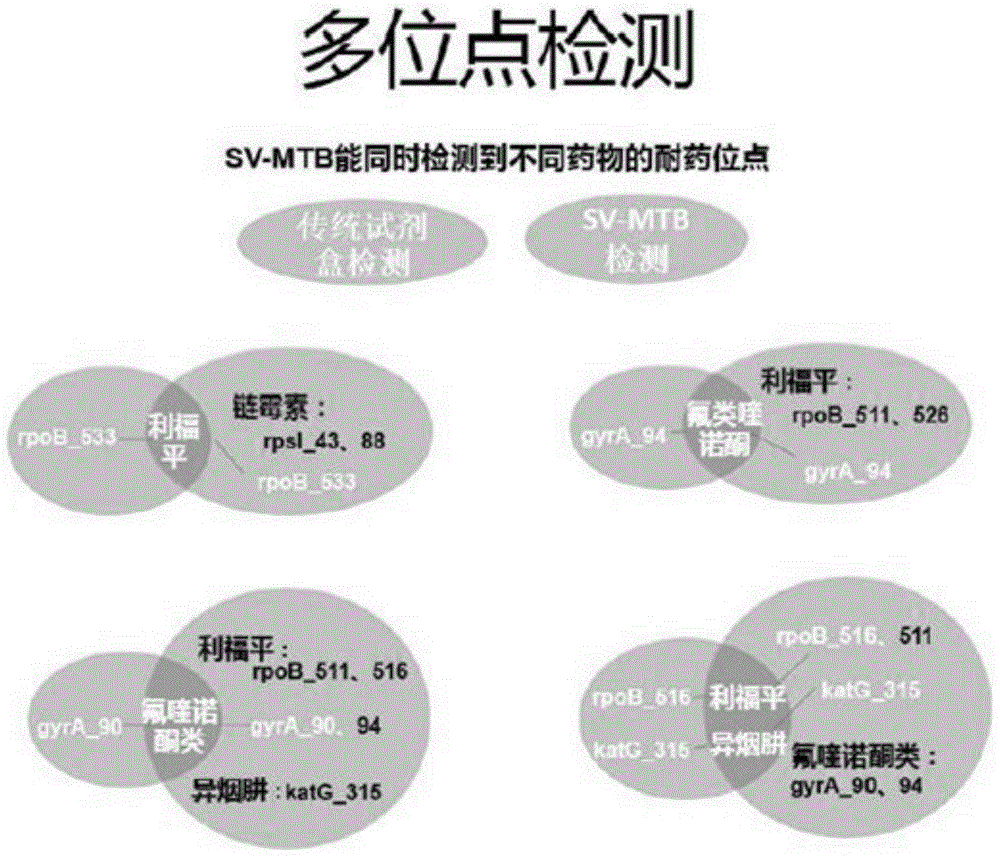
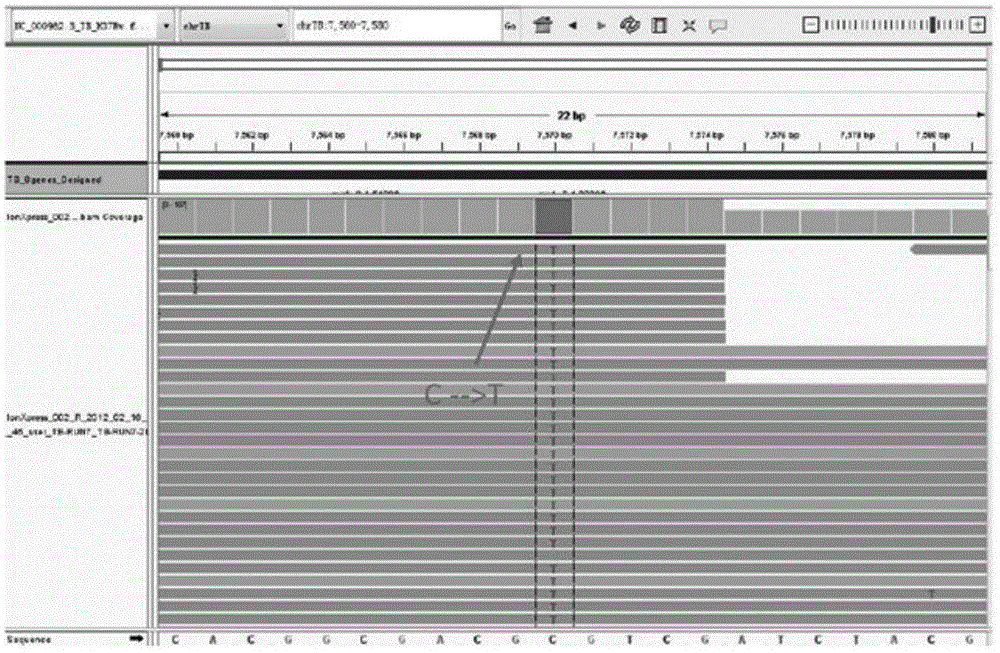
Method for detecting four tuberculosis rifampicin, isoniazide and fluoroquinolones-resistant genes
A fluoroquinolone and drug resistance gene technology, applied in the detection of four tuberculosis drug resistance genes of fluoroquinolone drugs, isoniazid, rifampicin, can solve the problem of inability to determine the nature of mutation, disease deterioration, scientific, safe, Accurately guide tuberculosis patients and other issues
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
[0053] In order to enable those skilled in the art to better understand the solution of the present invention, and to make the above-mentioned purpose, features and advantages of the present invention more obvious and understandable, the present invention will be further described in detail below in conjunction with the embodiments and the accompanying drawings of the embodiments.
[0054] The method for detecting four tuberculosis drug-resistant genes for rifampicin, isoniazid, and fluoroquinolones in Example 1 of the present invention includes the following steps:
[0055] 1. DNA quality inspection
[0056] 1) Detect DNA concentration with Qubit.
[0057] 2) For DNA with too low concentration, use agilent to detect whether the DNA is degraded by fragment size.
[0058] 3) To determine the 260 / 280 value of DNA, it is required to be greater than 1.6. In this example, the 260 / 280 value of DNA is greater than 1.6; / 280 value.
[0059] 2. Build the library
[0060] 1) Target ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com