DNA polymerase for improving synthesis efficiency of catalytic DNA
A technology of polymerase and ability, applied in the field of enzyme engineering, can solve the problem of short-term binding of Dbh to DNA
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0018] Example 1 Determination of Mutation Sites
[0019] By comparing homologous sequences, the non-conserved sites in the Dbh sequence and the mutation direction and frequency of these sites were determined. The results are shown in Table 1. Eight mutation directions of T37F, I62V, M76I, A221S, Y249I, L250V, K337R and KSKIP (241-245) RVRKS were determined, and the binding and binding of these eight mutants Dbh and DNA were simulated by computer. Combined free energy calculations. The results are shown in Table 2. In theory, the reduction of binding free energy means that the binding of Dbh to the substrate is more stable, which means that the affinity between the enzyme and the substrate is greater, so that the continuous synthesis ability of the enzyme is improved. From the analysis in Table 2, it can be seen that , all mutations except K337R and Y249I can enhance the processivity.
[0020] Table 1. Mutation residue types and frequencies of non-conservative amino acids
...
Embodiment 2
[0026] Embodiment 2 Sdbh mutant enzyme construction and processivity comparison
[0027] Eight mutants Dbh T37F, Dbh I62V, Dbh M76I, DbhA221S, DbhKSKIP(241-245)RVRKS, Dbh Y249I, Dbh L250V, Dbh K337R were constructed by site-directed mutagenesis, and the N-terminal of each mutant was connected to the protein by a flexible linker Sso7d, 8 corresponding Sdbh mutants were obtained: Sdbh T37F, Sdbh I62V, Sdbh M76I, Sdbh A221S, Sdbh KSKIP(241-245)RVRKS, Sdbh Y249I, Sdbh L250V, Sdbh K337R. The nucleotide sequence encoding Dbh is shown in SEQ ID NO.1, and the starting amino acid sequence of Dbh is shown in SEQ ID NO.2, which encodes Dbh after the N-terminus is connected to the protein Sso7d through a flexible linker, that is, the nucleotide of Sdbh The sequence is shown in SEQ ID NO.3, the nucleotide sequence encoding the flexible linker is shown in SEQ ID NO.4, and the nucleotide sequence encoding Sso7d is shown in SEQ ID NO.5.
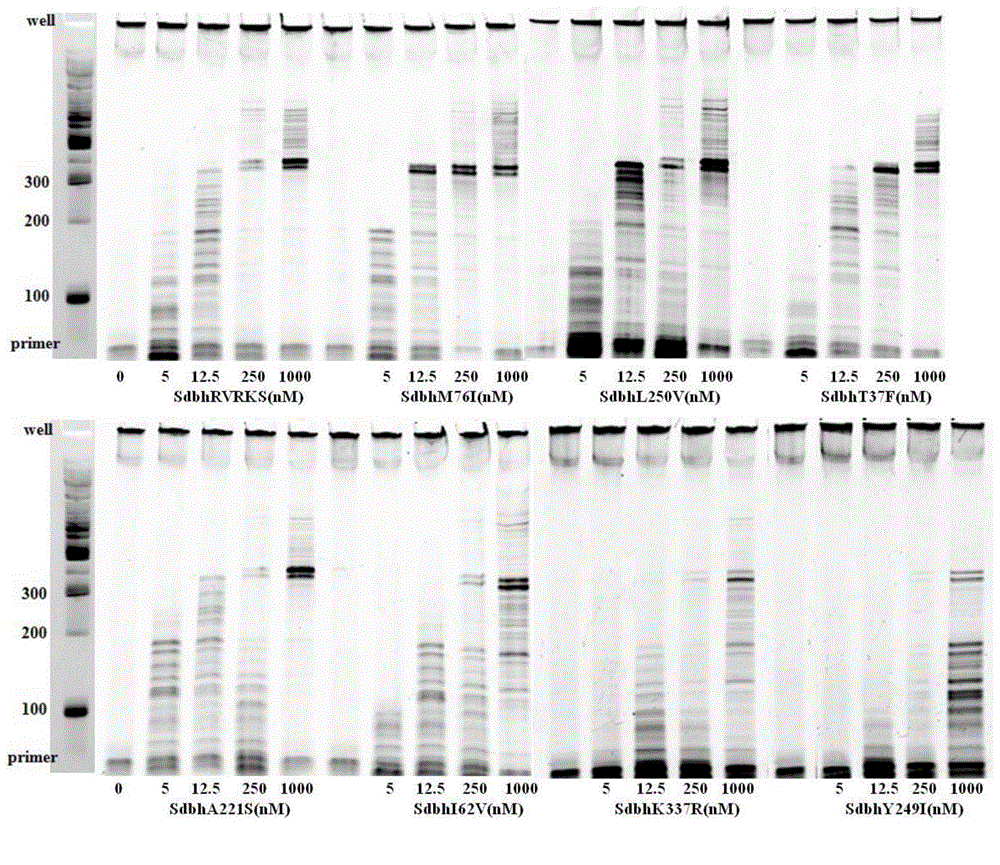
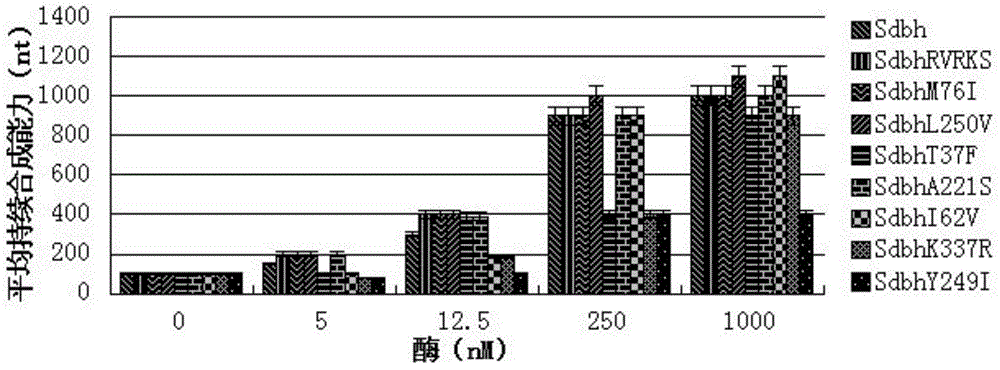
[0028] The processivity of Sdbh and its eight mutants...
Embodiment 3
[0031] The catalytic activity comparison of embodiment 3Sdbh and Sdbh mutant
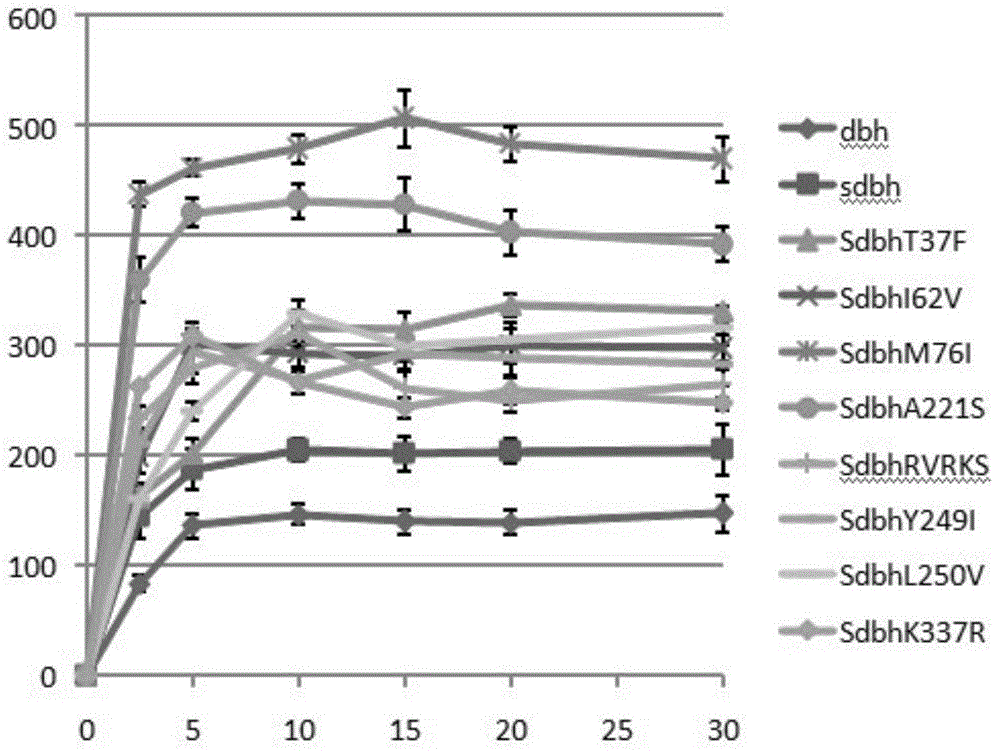
[0032] 12.5nM of the annealed primer / template was added to the reaction buffer (10mM HEPES NaOH (pH7.4), 50mMNaCl, 10mM MgCl 2, 200 mM dNTPs, 1 mM DTT, 100 μg / ml BSA and 0.1% Triton X-100). 12.5nM DNA polymerase was added to initiate DNA synthesis at 37°C. At different time points, 1 μL samples were added to 99 μL PicoGreen (Molecular Probes) diluted 1:200, and reacted in TE buffer (10 mM Tris-Hcl pH 8.0 and 1 mM EDTA). The amount of synthesized DNA was quantified using an H1 hybrid multi-function microplate reader (Burten Instruments, USA). The unit activity of DNA polymerases was determined by comparing their initial rates with Sdbh.
[0033] The result is as figure 2 As shown, the Sdbh mutant exhibited an increase in polymerase activity compared to Sdbh, suggesting that mutations of non-conserved residues do not reduce polymerase activity but significantly increase the initial rate of DNA sy...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com