A method for rapidly evaluating the resistance of rice srbsdv varieties by detecting the virus proliferation rate in the coleoptile
A virus multiplication and coleoptile technology, applied in the field of agricultural science, can solve problems such as low accuracy and long identification period
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment
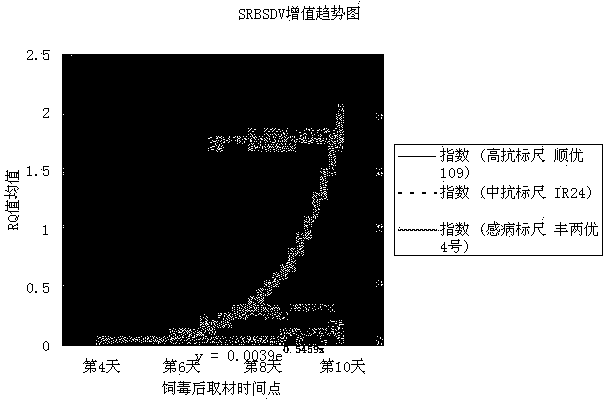
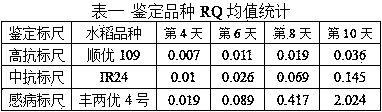
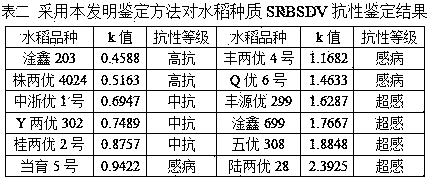
[0050] In 2015, the identification method of this invention was used to conduct SRBSDV resistance identification on some rice varieties resources collected. Three rice varieties, Shunyou 109, IR24, and Fengliangyou 4 were set as the identification scales for high resistance, medium resistance, and susceptibility. Among them, Shunyou 109 had a disease resistance performance of 138 in the 2013-2014 indoor inoculation identification. Among the rice variety resource groups, it ranks 11, IR24 ranks 28, and Fengliangyou 4 ranks 69. Coleoptile collection time points were set to four time points on the 4th, 6th, 8th, and 10th days after the completion of inoculation, and 3 replicates were taken for each time point for each species.
[0051] Real-time fluorescent quantitative PCR obtains the RQ value of each species' periodic sampling time point, as shown in Table 1:
[0052]
[0053] Then draw the trend line of the SRBSDV proliferation index: take the average RQ value as the ordinate, and...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com