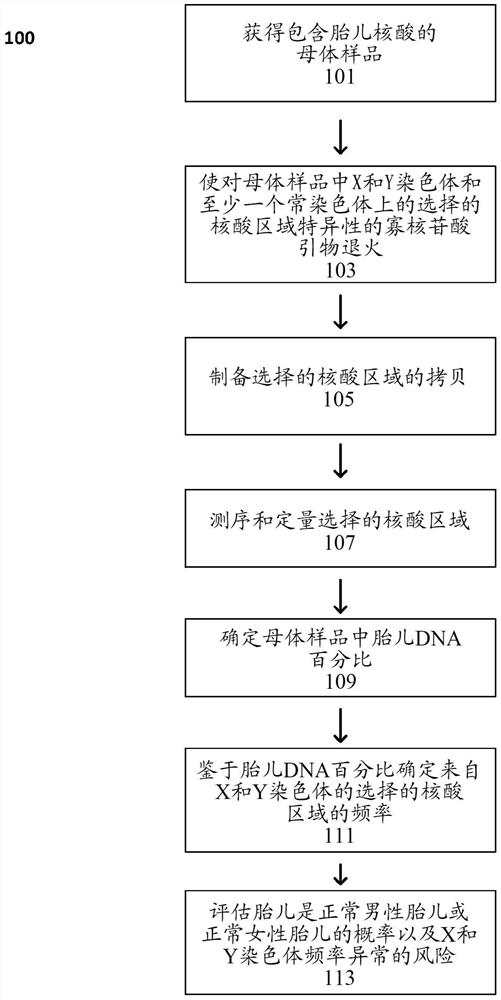
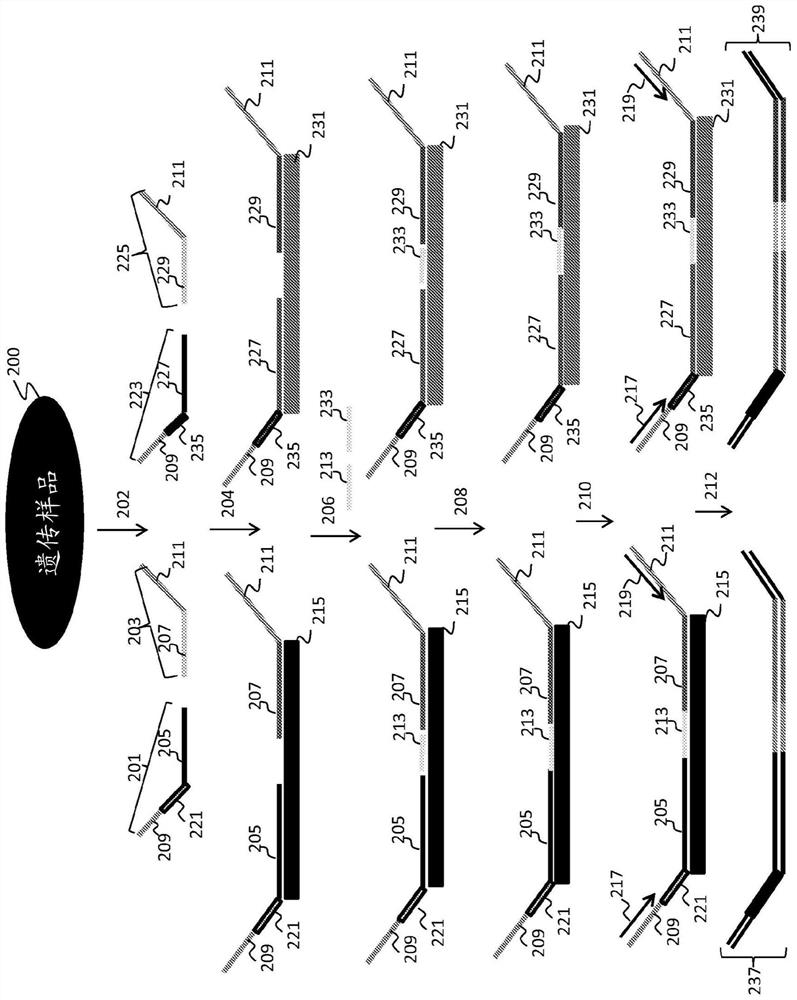
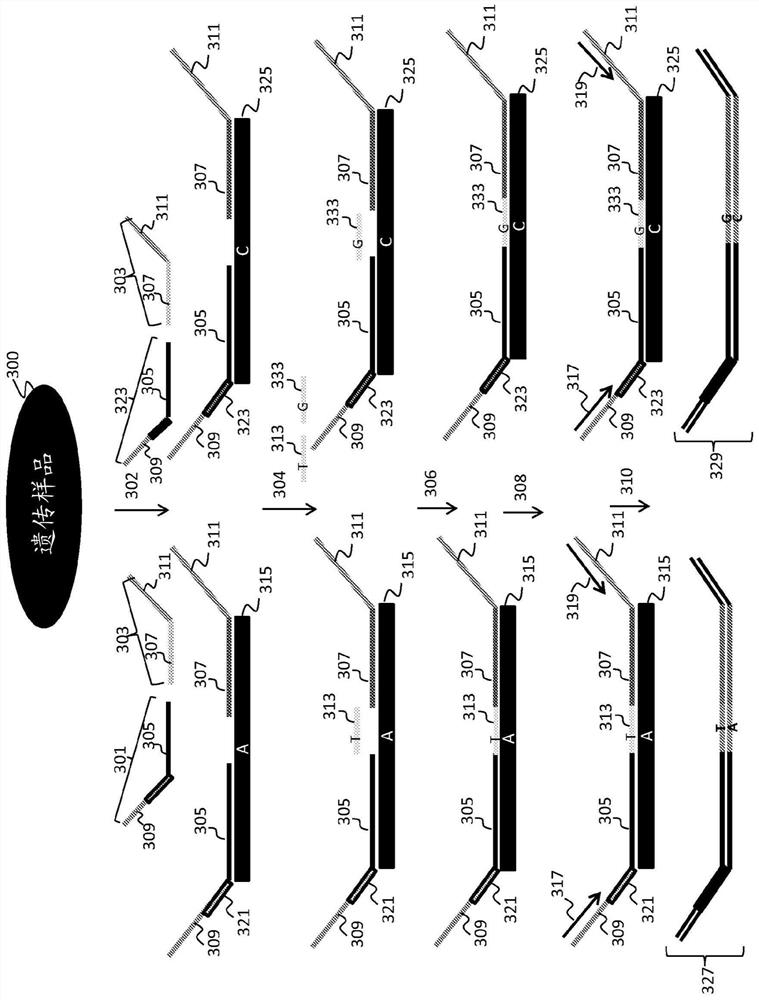
Statistical analysis for noninvasive chromosomal aneuploidy determination
An aneuploidy, X-chromosome technique used in the field of statistical analysis for non-invasive sex chromosome aneuploidy determination
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0232] Example 1: Preparation of DNA for the Tandem Ligation Procedure
[0233] Genomic DNA from subjects was obtained from Coriell Cell Repositories (Camden, New Jersey) and fragmented by acoustic shearing (Covaris, Woburn, MA) to an average fragment size of approximately 200 bp.
[0234] DNA was biotinylated using standard procedures. Briefly, Covaris fragmented DNA ends were repaired by generating the following reactions in 1.5 ml microtubes. 5ug DNA, 12 μl 10x T4 ligase buffer (Enzymatics, Beverly MA), 50 U T4 polynucleotide kinase (Enzymatics, Beverly MA) and H 2 0 to 120 μl. The microtubes were incubated at 37°C for 30 minutes. DNA was diluted with 10 mM Tris 1 mM EDTA pH 8.5 to a desired final concentration of ~0.5 ng / μl.
[0235] 5 μl of DNA was placed in each well of a 96-well plate, and the plate was sealed with adhesive parafilm and spun at 250×g for 10 seconds. The plate was then incubated at 95°C for 3 minutes, cooled to 25°C, and spun again at 250 xg for 10 ...
Embodiment 2
[0241] Example 2: Universal amplification of ligated products
[0242] The polymerized and / ligated nucleic acid is amplified using universal PCR primers complementary to the universal sequence present in the first and second fixed sequence oligonucleotides that hybridize to the nucleic acid region of interest. 25 μl of each of the reaction mixtures in Example 3 were used in each amplification reaction. 50 μL Universal PCR consisting of 25 μL of eluted ligation product plus 1x Pfusion buffer (Finnzymes, Finland), 1 M betaine, 400 nM of each dNTP, 1 U Pfusion error-correcting thermostable DNA polymerase, and primer pairs with sample tags Reactions are used to uniquely identify individual samples prior to pooling and sequencing. Using BioRad Tetrad TM Thermal cyclers perform PCR under stringent conditions.
[0243] 10 μl of universal PCR products from each of the samples were pooled and quantified using Quant-iT TM PicoGreen, (Invitrogen, Carlsbad, CA) purified and quantifie...
Embodiment 3
[0244] Example 3: Analysis of polymorphic loci to assess percent fetal contribution
[0245] To assess the proportion of fetal nucleic acid in a maternal sample, an assay was designed for a panel of SNP-containing loci on chromosomes 1 to 12, in which two bridging oligonucleotides differing by one base were used to interrogate each SNP (see, e.g. image 3 ). SNPs were optimized for minor allele frequency in the HapMap 3 dataset. Duan, et al., Bioinformation, 3(3):139-41 (2008); Epub 2008 Nov 9.
[0246] Oligonucleotides were synthesized by IDT (Coralville, Iowa) and pooled together to generate a single multiplexed assay pool. PCR products were generated from each subject sample as previously described. An informative polymorphic locus is defined as a locus in which the fetal allele differs from the maternal allele. Because the assay exhibits an allele specificity of over 99%, informative loci are readily identified when the fetal allelic ratio of the locus is measured to b...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com