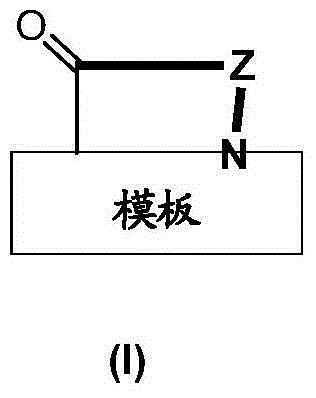
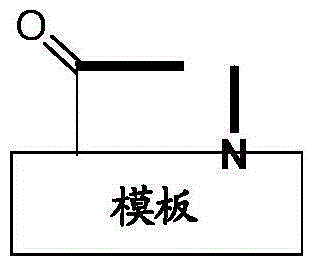
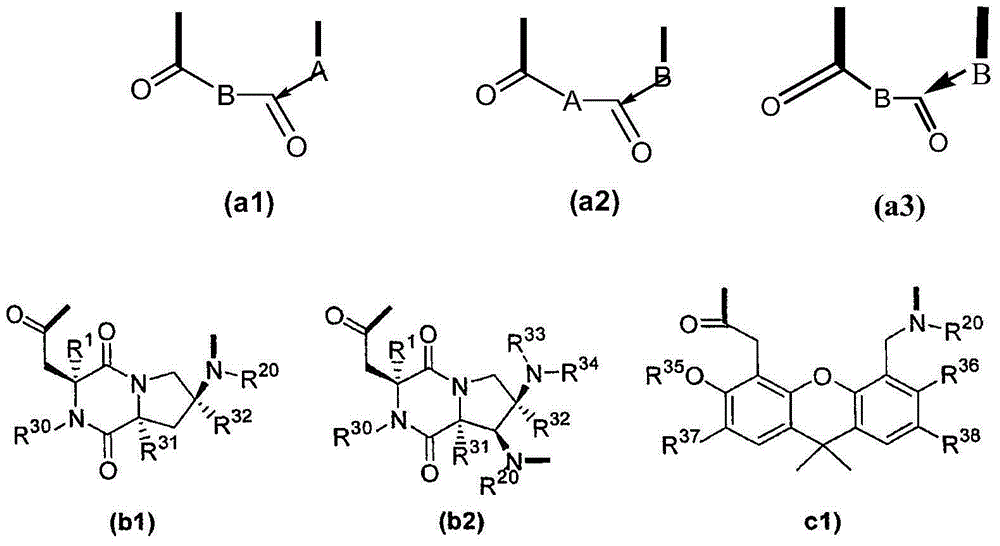
Template-fixed beta-hairpin peptidomimetics with protease inhibitory activity
A definition, compound technology, applied in the field of template-immobilized β-hairpin peptidomimetics with protease inhibitory activity, capable of solving the problem of not exhibiting high selectivity and particularly high potency
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0595] Example 1 (15.37), Example 2 (11.54), Example 3 (7.82), Example 4 (8.62), Example 5 (16.51), Example 6 (13.67), Example 7 (3.61), implementation Example 8 (4.11), Example 9 (5.82), Example 10 (7.98), Example 11 (8.38), Example 12 (6.80), Example 13 (7.41), Example 14 (6.20), Example 15 (8.68), Example 16 (9.82), Example 17 (5.59), Example 20 (7.32), Example 21 (8.66), Example 22 (8.68), Example 23 (12.66), Example 24 (8.67), Example 25 (7.53), Example 26 (9.02), Example 27 (8.06), Example 28 (9.62), Example 29 (8.78), Example 30 (10.49), Example 31 ( 5.50), Example 32 (7.45), Example 33 (8.39), Example 34 (10.16), Example 35 (9.04), Example 36 (10.98), Example 37 (7.56), Example 38 (9.29 ), Example 39 (8.32), Example 40 (10.11), Example 41 (7.23), Example 42 (8.83), Example 43 (7.92), Example 44 (9.87), Example 45 (8.26) , Example 52 (6.20), Example 53 (8.68), Ex54 (9.82), Example 55 (5.59), Example 56 (6.06), Example 57 (6.47), Example 58 (7.32), Example 59 (8.68), ...
Embodiment 46
[0596] Example 46 is shown in Table 1. Peptides were synthesized starting from the amino acid Pro grafted to the resin. The starting resin was Fmoc-Pro-2-chlorotrityl resin, which was prepared as described above. According to the procedure described above, a linear peptide was synthesized on a solid support with the following sequence: Resin-Pro- D Asp(OtBu)-P11-P10-P9-P8-P7-P6-P5-P4-P3-P2-P1. Afterwards, disulfide bond formation was followed as described in Procedure B, and the peptide was cleaved from the resin, cyclized, deprotected, and purified.
[0597] Using the analytical method described above, determine the HPLC-retention time (minutes):
[0598] Example 46 (8.94).
Embodiment 47
[0599] Example 47 is shown in Table 1. Peptide synthesis begins with the amino acid Asp grafted to the resin. The starting resin was Fmoc-Asp(OtBu)-2-chlorotrityl resin, which was prepared as described above. According to the procedure described above, a linear peptide was synthesized on a solid support with the following sequence: Resin-Asp(OtBu)- D Pro-P11-P10-P9-P8-P7-P6-P5-P4-P3-P2-P1. Afterwards, disulfide bond formation was followed as described in Procedure B, and the peptide was cleaved from the resin, cyclized, deprotected, and purified.
[0600] Using the analytical method described above, determine the HPLC-retention time (minutes):
[0601] Example 47(7.29).
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com