High-throughput retrieval method for drug targets
A high-throughput, drug-based technology, applied in the field of bioinformatics, can solve difficult problems such as protein database structure screening, and achieve the effect of accelerated retrieval
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
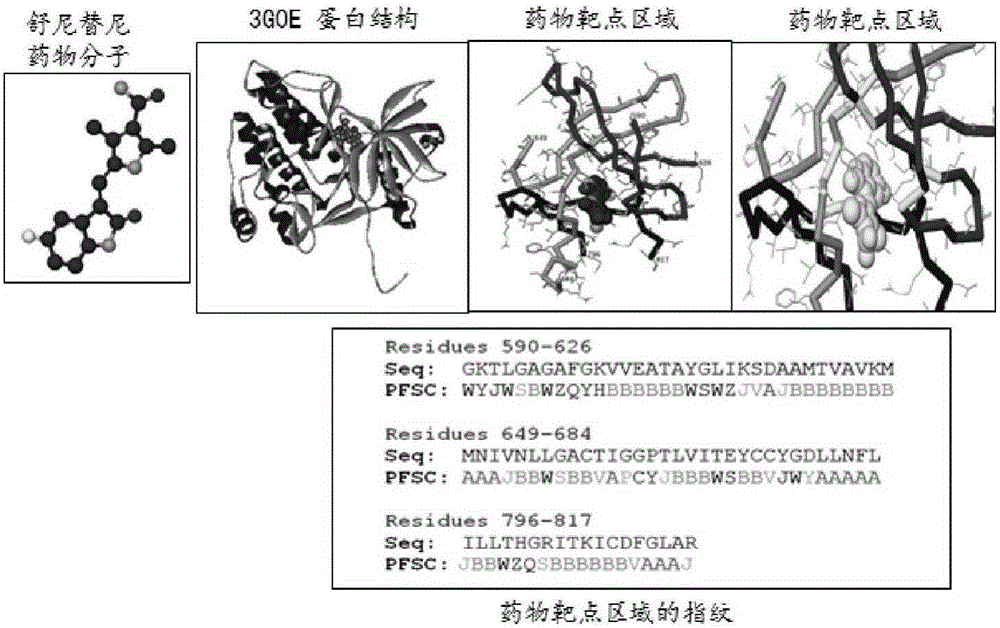
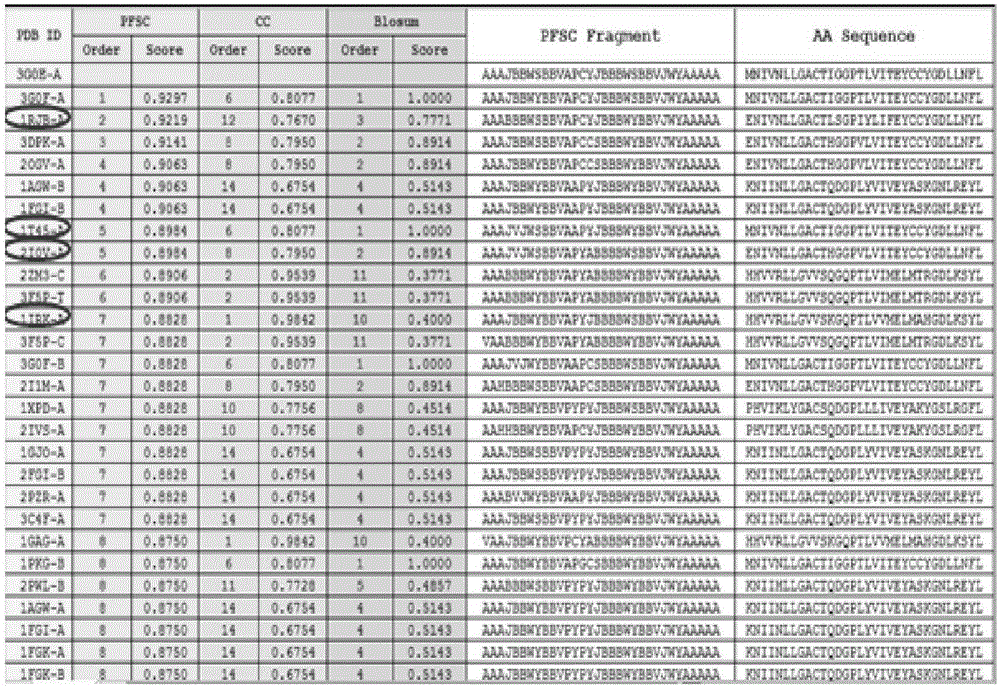
[0039] The method of the invention can be applied to the research direction of the secondary development of medicines, and for the approved clinical medicines, new efficacy can be developed by discovering new targets. In addition, it is applied to the development of protein probe molecules for diseases.
[0040] Case studies have been carried out for several drug molecules, each representing different initial data conditions in the database. Using the knowledge of the drug database and other databases for comparative verification, it was found that most of the protein targets with higher fingerprint scores were basically the same. But at the same time, new protein targets are also discovered. These new targeted proteins may lead to the identification of drug mechanisms and application to new therapeutic areas, expanding the indications of drugs. Here, the anticancer drug molecule sunitinib is taken as an example to illustrate how to discover the multiprotein target of the dru...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com