PCR detection primer and method for anoectochilus roxburghii colletotrichum orbiculare
A technology for anthracnose bacteria and detection primers, which is applied in biochemical equipment and methods, microbial determination/inspection, DNA/RNA fragments, etc., can solve the problems of time-consuming, inability to achieve rapid inspection, etc., and achieves simple, fast and practical operation. Good and practical effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0030] Example 1: Primers specific amplification of Clematis anthracnose bacteria
[0031] 1. Specific Detection of Anthracnose Bacteria in Clematis aureus
[0032] PCR reaction system 20.0μL, including 2× Taq PCRMasterMix 10.0 μL, primers CR1 and CF1 5 μM each, template DNA 50-100 μg, d.d.H 2O to make up to 20.0 μL. The PCR reaction conditions were: pre-denaturation at 95°C for 5 min, denaturation at 95°C for 30 sec, annealing at 65°C for 30 sec, extension at 72°C for 30 sec, a total of 35 cycles, and extension at 72°C for 10 min.
[0033] 2. Test results
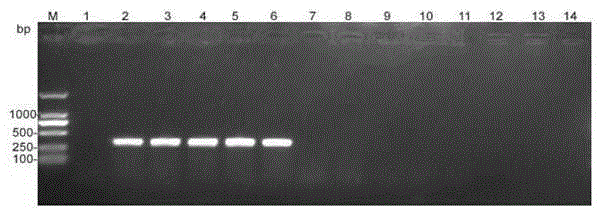
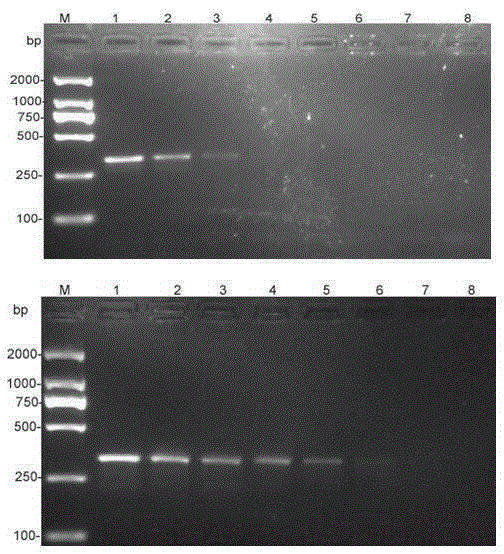
[0034] Specificity of detection: Except for the 326bp product that can be specifically amplified from the DNA of Clematis anthracnose, the DNA of other 21 different fungi failed to amplify any product, which has strong specificity.
Embodiment 2
[0035] Embodiment 2: Sensitivity detection of primers to Clematis anthracnose bacteria
[0036] 1. Dilution of DNA concentration: the extracted genome DNA of Clematis anthracis was measured by a spectrophotometer, and then diluted in series.
[0037] 2. Sensitivity Detection of Anthracnose Bacteria from Clematis aureus
[0038] PCR reaction system 20.0μL, including 2× Taq PCRMasterMix 10.0 μL, primers CR1 and CF1 5 μM each, template DNA 50-100 μg, d.d.H 2 O to make up to 20.0 μL. The PCR reaction conditions were: pre-denaturation at 95°C for 5 min, denaturation at 95°C for 30 sec, annealing at 65°C for 30 sec, extension at 72°C for 30 sec, a total of 35 cycles, and extension at 72°C for 10 min.
[0039] 3. Detection results: In the 20.0μL reaction system, the genomic DNA of Clematis anthracis can obtain obvious amplification bands, and the detection sensitivity can reach 11.2×10 -5 ng / μL.
Embodiment 3
[0040] Example 3: Detection of Clematis anthracnose in diseased plant samples.
[0041] 1. Sample collection: Clematis plant tissue samples were collected from the Clematis production base in Yongtai County, Fuzhou City, Fujian Province.
[0042] 2. DNA extraction and detection
[0043] DNA was extracted from diseased plant tissue using the NaoH rapid lysis method, and PCR amplification was performed according to the above method. The PCR reaction system was 20.0 μL, including 2× Taq PCRMasterMix 10.0 μL, primers CR1 and CF1 5 μM each, template DNA 50-100 μg, d.d.H 2 O to make up to 20.0 μL. The PCR reaction conditions were: pre-denaturation at 95°C for 5 min, denaturation at 95°C for 30 sec, annealing at 65°C for 30 sec, extension at 72°C for 30 sec, a total of 35 cycles, and extension at 72°C for 10 min.
[0044] 3. Test results
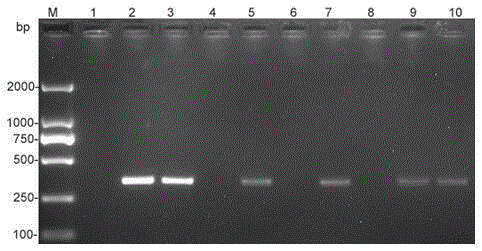
[0045] see results image 3 , if the 326bp product can be specifically amplified, it can be judged that there is Clematis anthracis in ...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Sensitivity | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com