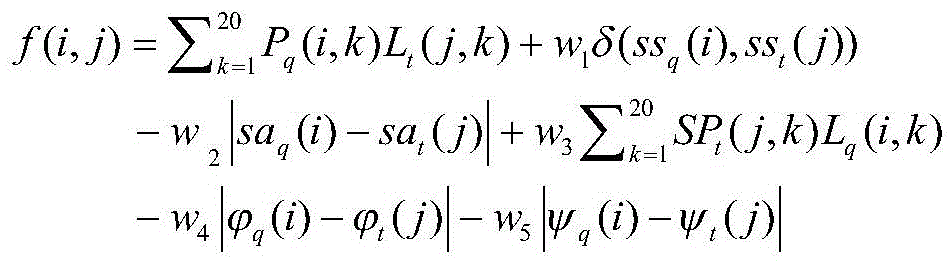Method for constructing distance model between protein residues based on Bolzmann probability density function
A technology of probability density function and construction method, which is applied in the field of computer application and bioinformatics, can solve the problems of low precision and weak sampling ability of conformational space, and achieve the effect of high precision and strong sampling ability of conformational space
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
[0042] The present invention will be further described below in conjunction with the accompanying drawings.
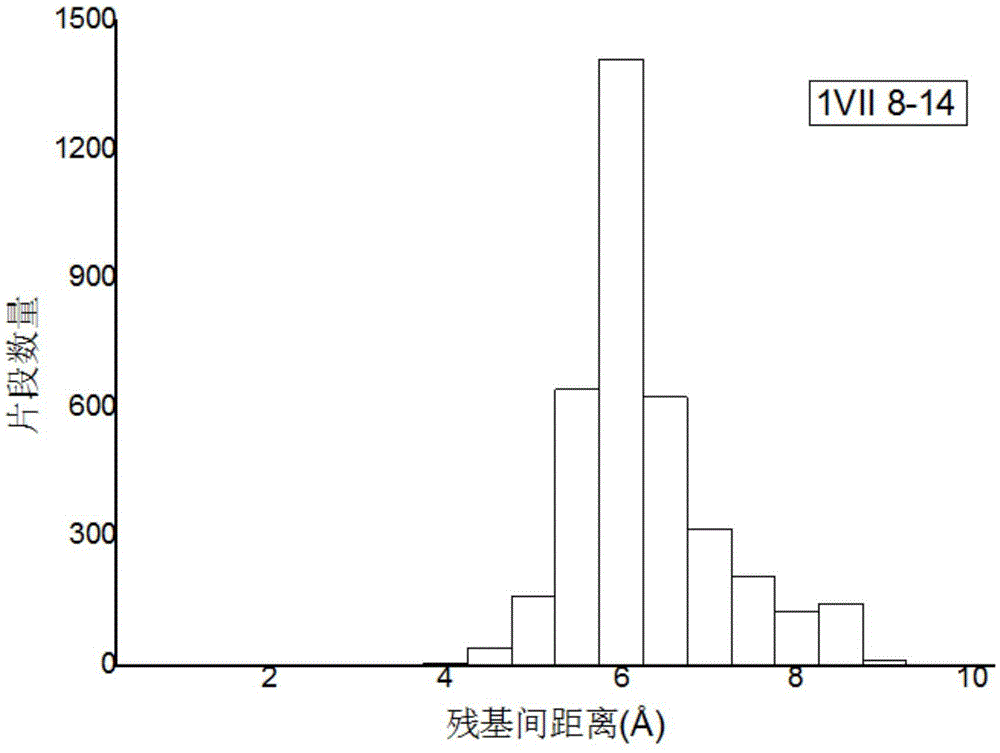
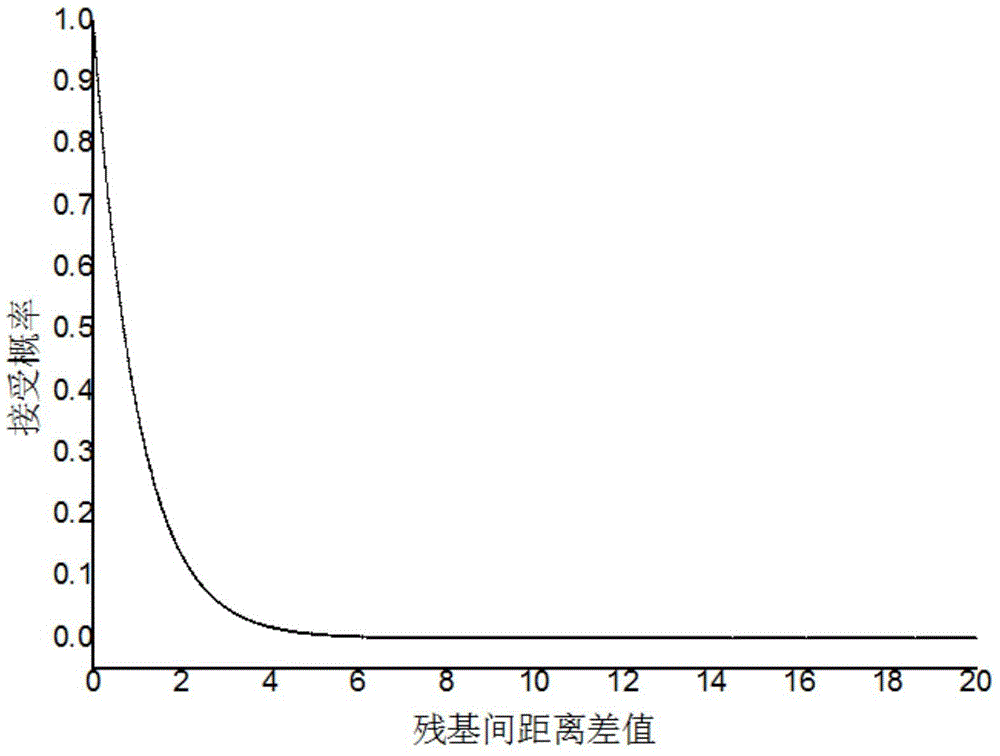
[0043] refer to figure 1 with figure 2 , a method for constructing probability density function constraints based on the distance between protein residues, comprising the following steps:
[0044] 1) Build a non-redundant template library;
[0045] 1.1) Download the resolution less than from the protein database (PDB) website high-precision protein, where is the distance unit,
[0046] 1.2) Split the protein containing multiple polypeptide chains into single chains, and keep the longest chain to compare sequence similarity with other chains, and remove redundant polypeptide chains with a similarity greater than 30%;
[0047] 1.3) Calculate the sequence similarity I of the remaining polypeptide chains in pairs mn , to count the cumulative similarity of each chain Wherein m and n are the serial numbers of the polypeptide chain, and N is the total number of al...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com