New fusion gene detected in lung cancer
A detected and gene fusion technology, applied in fusion polypeptide, genetic engineering, detection of programmed cell death, etc., can solve problems such as differences in drug reactivity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0455] This example describes the identification of novel fusion transcripts in lung cancer tissue.
[0456] To identify novel fusion transcripts that could serve as therapeutic targets, whole-transcriptome sequencing of 114 lung adenocarcinomas and 3 noncancerous lung tissues was performed (RNA sequencing, Meyerson, M. et al., NatRev Genet, 2010, 11, 685-696 ).
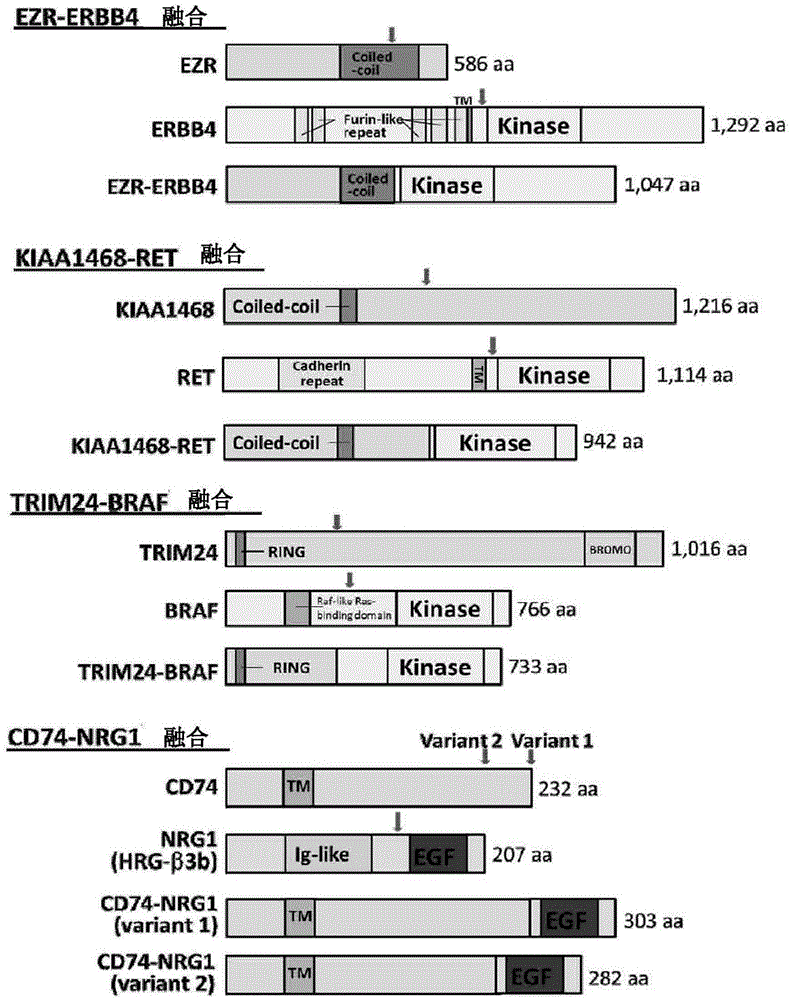
[0457] Analyze the Paired-endRead obtained by RNA sequencing, and perform Sanger sequencing of reverse transcription (RT)-PCR products. The results are shown in Table 2 and figure 1 As shown, four novel fusion gene products were identified.
[0458] [Table 2]
[0459]
[0460] EZR-ERBB4 is produced by chromosomal transposition t(2;6), and is a fusion gene of the EZR gene present on chromosome 6q25 and the ERBB4 gene present on chromosome 2q34.
[0461] KIAA1468-RET is produced by chromosomal transposition t(10;18), and is a fusion gene of the KIAA1468 gene present on chromosome 18q21 and the RET gene present o...
Embodiment 2
[0466] This example describes the RT-PCR detection of the gene fusions discovered in Example 1.
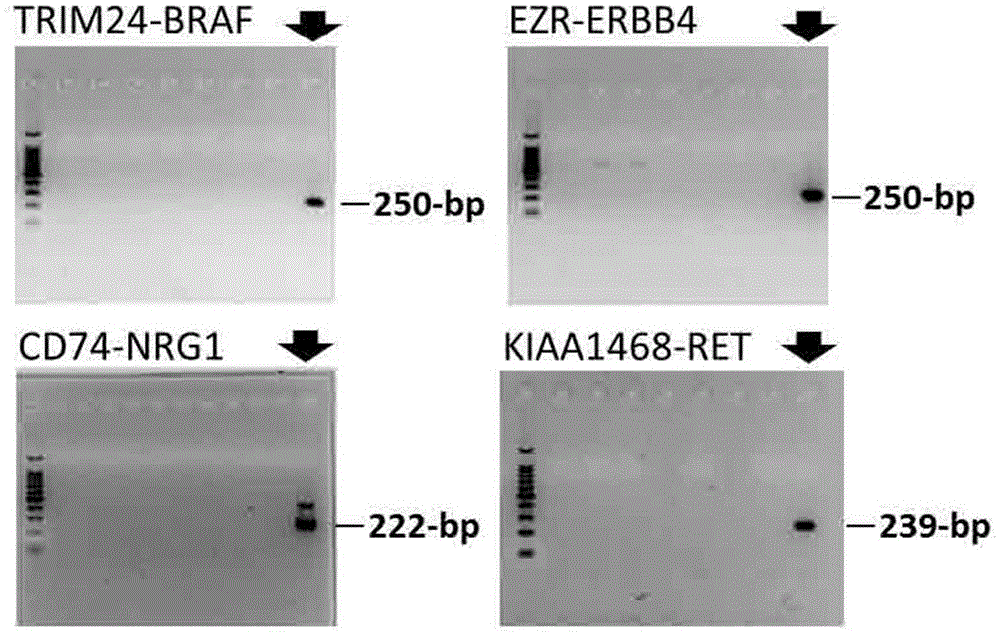
[0467] For each fusion gene, PCR primers (forward primer and reverse primer, respectively) derived from the cDNA sequences of the 5' side and 3' side gene parts were prepared (Table 1). Using these primers, PCR amplification was performed using cDNA synthesized from cancer tissue-derived RNA as a template.
[0468] exist figure 2 Electropherogram showing PCR products. Amplification of a specific band was confirmed in a part of the samples, and the base sequence of the PCR product was further confirmed. As a result, it was confirmed that a part of the fusion gene was amplified.
[0469] From the above results, it was shown that the gene fusion found in Example 1 could be detected by checking whether the amplification of a specific band was obtained by RT-PCR and determining the base sequence of the PCR product.
Embodiment 3
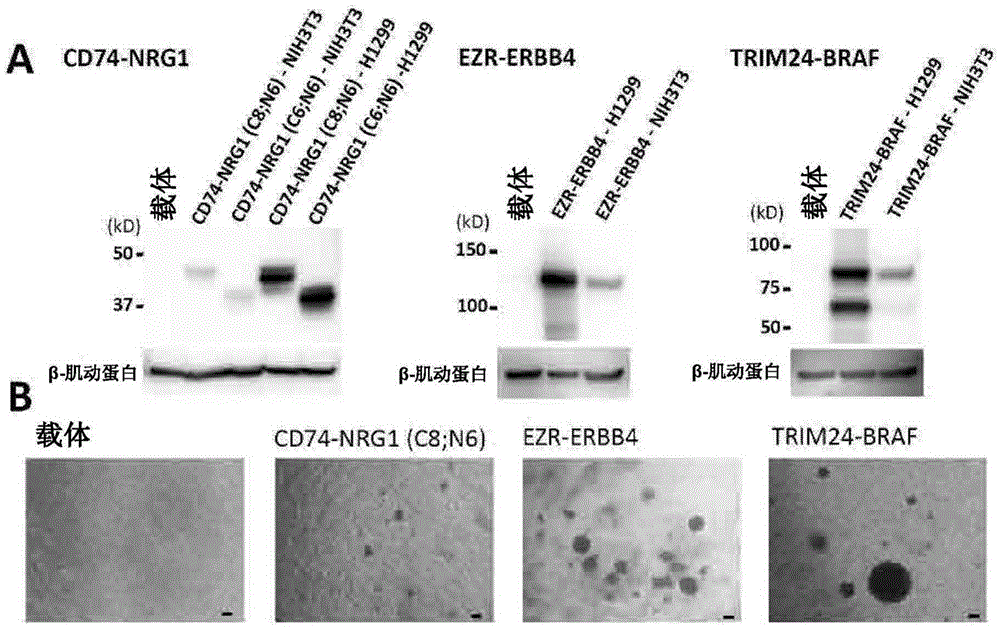
[0471]This example shows that the gene fusion found in Example 1 is highly likely to be the responsible mutation for lung cancer.
[0472] In the five cases of lung cancer in which novel gene fusions were found in Example 1, whether there were EGFR point mutations-in-frame deletion mutations (EGFR point mutations, EGFRin-flamedeletion mutations), KRAS point mutations (KRAS point mutations), BRAF mutations, etc. Point mutation (BRAF pointmutation), HER2 in-frame insertion mutation (HER2in-flameinsertionmutation), EML4-ALK gene fusion (EML4-ALKfusion), KIF5B-RET gene fusion (KIF5B-RETfusion), CCDC6-RET gene fusion (CCDC6-RETfusion), CD74-ROS1 gene fusion (CD74-ROS1fusion), EZR-ROS1 gene fusion (EZR-ROS1fusion) and SLC34A2-ROS1 gene fusion (SLC34A2-ROS1fusion).
[0473] As a result, all of the above-mentioned 5 cases of lung cancer were negative for other known mutations, and the 4 novel gene fusions were mutually exclusive with other known mutations responsible for cancer.
[0...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com