Exome potential pathogenic mutation detection method based on family line
A technology for exome sequencing and mutation detection, which is applied in biochemical equipment and methods, microbial measurement/testing, instruments, etc., and can solve problems such as high heterogeneity, pathogenic variant mining of sequencing data, and high mutation rate
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
[0031] It should be noted that, in the case of no conflict, the embodiments of the present invention and the features in the embodiments can be combined with each other.
[0032] The present invention will be described in detail below with reference to the accompanying drawings and examples.
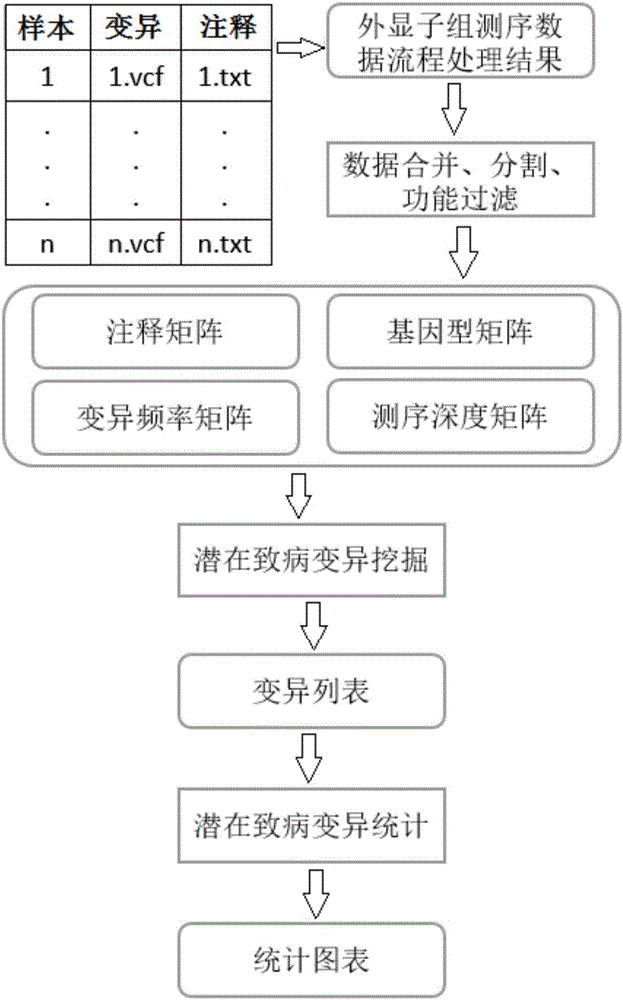
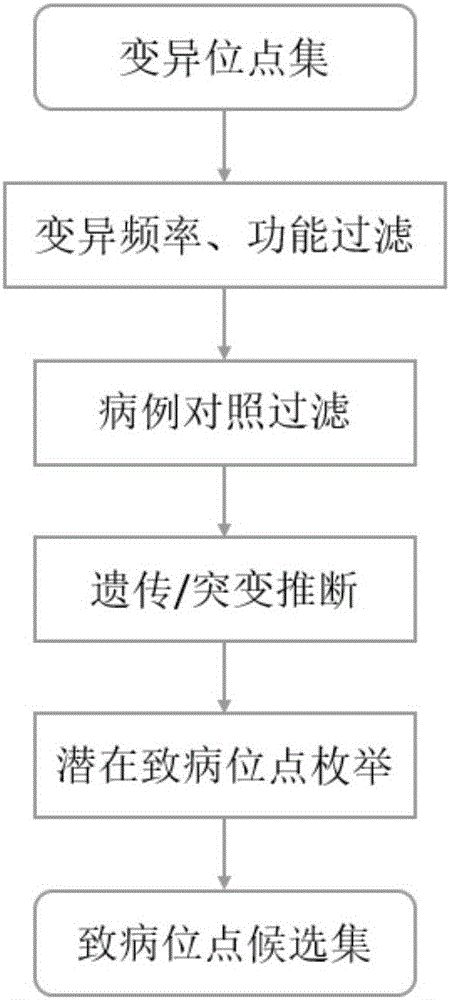
[0033] Such as figure 1 Shown, the present invention mainly comprises 4 steps:
[0034] (1) Read the result file of the exome sequencing data processing flow, merge the variation vcf file and ANNOVAR annotation file of each sample, and perform functional filtering to obtain a preliminary integration file. Each sample is divided into SNP and SNP according to the variation type. INDEL two files;
[0035] Among them, the ANNOVAR annotation result file must be input from the vcf file corresponding to the name, and obtained by using ANNOVAR annotation. It is a table, each row records a variation, and each column records an annotation, including and only including the following columns:
[...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com