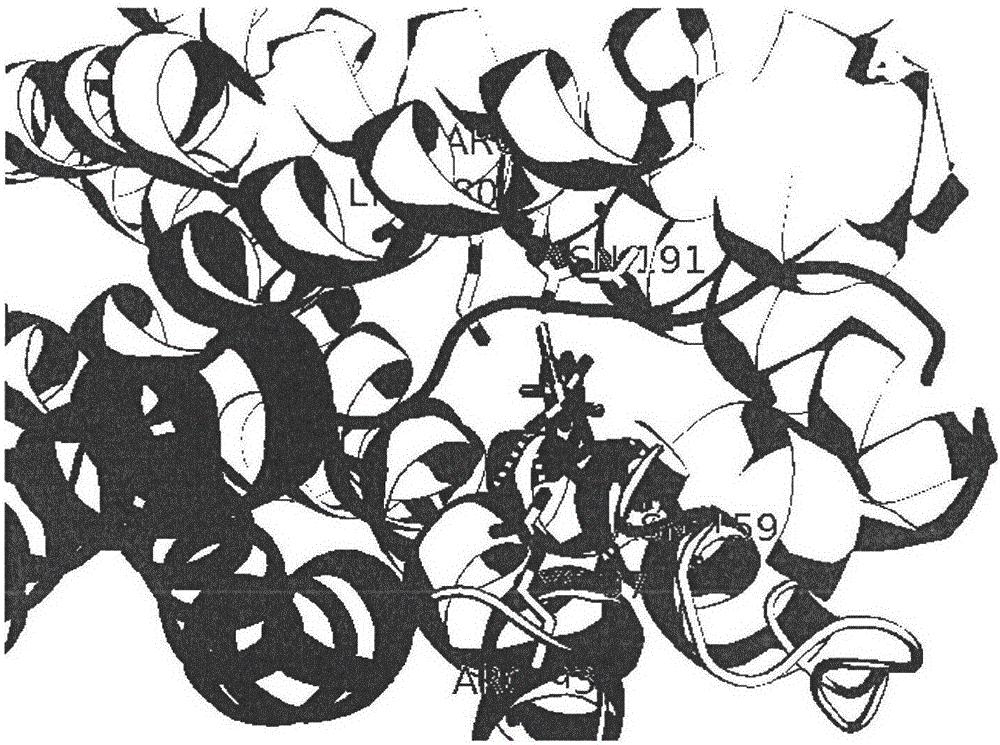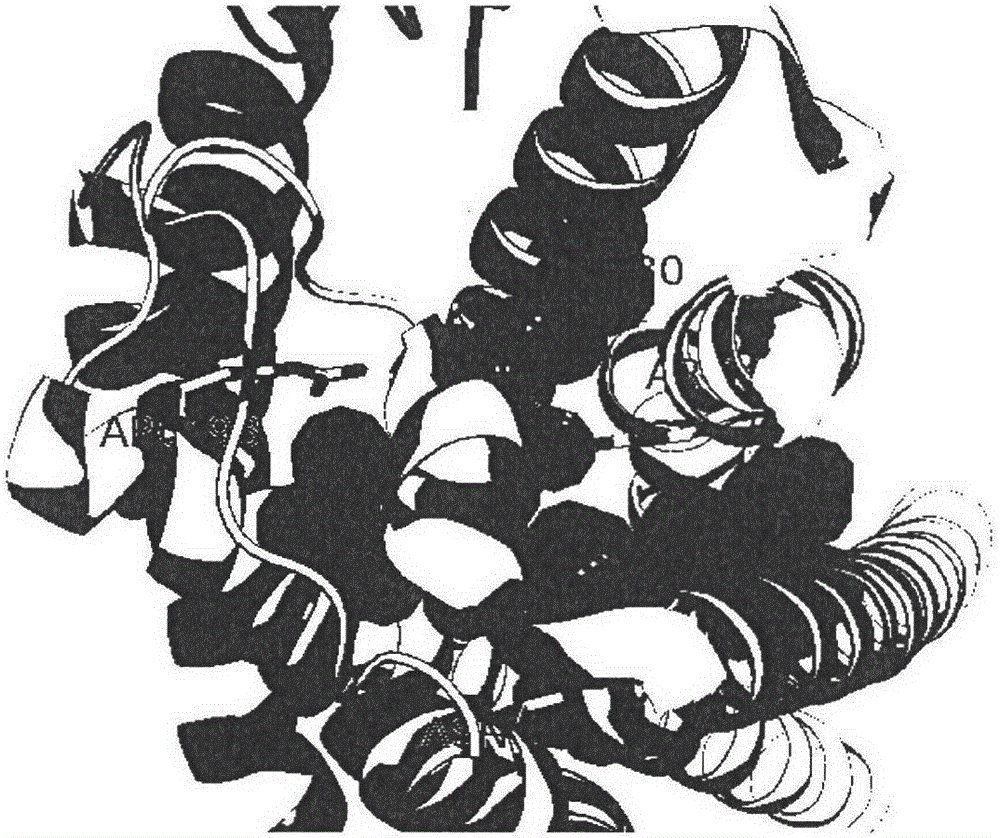Computer drug design method using P2Y12 as target and application thereof
A technology of P2Y12 and design method, applied in computing, special data processing applications, instruments, etc., can solve problems such as slow research progress and restrictions on the development of antithrombotic drugs
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
[0027] see Figure 1 to Figure 6 Binding pockets were identified for P2Y12 using the Discovery Studio program as shown.
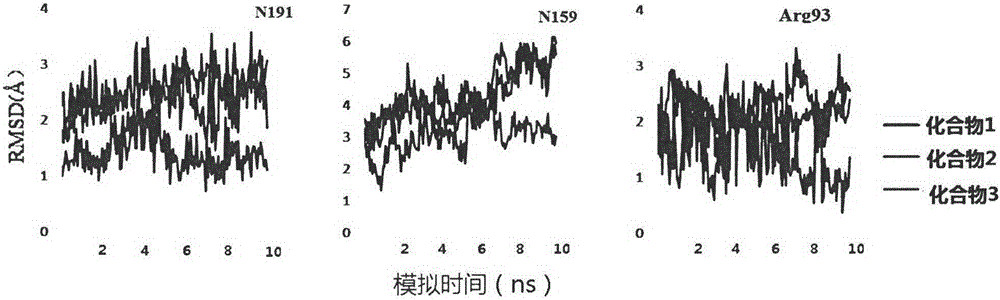
[0028] Load the acceptor PDB structure to display the PDB structure in cartoon mode. The main residues that small molecules interact with receptors are N191, N159, Arg93, Cys97 residues.
[0029] (2) Using Autodock software to conduct virtual screening of the small molecule database of P2Y12 inhibitors. The crystal structure after homology modeling was processed with Autodock Tools, redundant molecules were removed, molecular optimization was performed according to Autodock Vina operation rules, charges were added, and the acceptor structure was converted into pdbqt format. All compound structures were downloaded from the Chembridge database, processed with PyRx and converted into pdbqt format. A processed compound database was built. Binding pocket regions were defined and the pocket radius was defined as 5 angstroms.
[0030] (3) According to the ident...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com