Method for diagnosing colorectal cancer from human feces sample by quantitive pcr, primers and kit
A technology for colorectal cancer and stool samples, which is applied in the direction of biochemical equipment and methods, and microbial measurement/testing, and can solve the problems of industrial differences in microbial groups and high costs
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0181] Example 1.- Materials and methods
[0182] 1. Biological samples
[0183] Stool samples were obtained from 27 patients with recently diagnosed colorectal cancer (CRC) (stages 0-I), 24 Lynch syndrome carriers (L) and 19 healthy individuals (C). All patients underwent colonoscopy at least 15 days before sample collection.
[0184] All personnel signed the corresponding informed consent. Exclusion criteria included antibiotic treatment within 1 month before the study and age <18 years.
[0185] 2.16S rDNA bacterial sequence:
[0186] B3, B10, B41, B46, B48 and B50 sequences correspond to denaturing gradient gel electrophoresis (DGGE) gel bands previously isolated from uncultured bacterial isolates.
[0187] -16S rDNA bacterial sequence SEQ ID NO: 1 (B3), published as GQ411111.1 in the EMBL-EBI European Nucleotide Archive (ENA) database;
[0188] -16S rDNA bacterial sequence SEQ ID NO: 4 (B10), published as GQ411118.1 in the EMBL-EBI European Nucleotide Archive (ENA)...
Embodiment 2
[0216] Example 2: Primer Design and Verification
[0217] Primers for the quantification of bacterial markers B3, B10, B46, B48, B41 and B50 in biopsies were designed based on comparative analysis with sequences previously obtained from phylogenetic groups using the following bioinformatic tools: Bioinformatics from Europe ClustalX (European Bioinformatics Institute (www.ebi.ac.uk)), Netprimer ((Premier Biosoft) and PrimerExpress (Life Tecnologies-Thermo Fischer).
[0218] The detection system chosen is Primer sets with less than 3 different positions relative to adjacent sets were discarded, as were primer sets with Tm values in dissociation curves different from the expected value. Primer validation was performed in biopsy samples and stool samples. Dissociation curves were measured to analyze primer performance.
[0219] exist Figures 8a to 13b In , the amplification curves and dissociation curves of the primers designed to amplify B3, B10, B46, B48, B41 and B50 a...
Embodiment 3
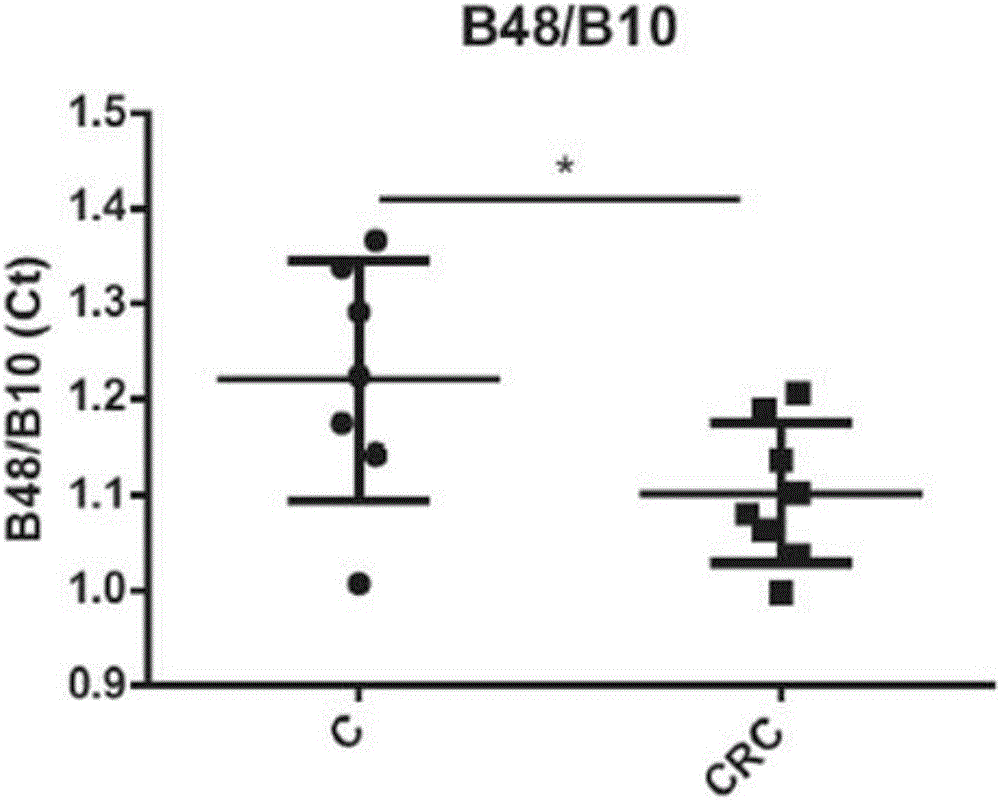
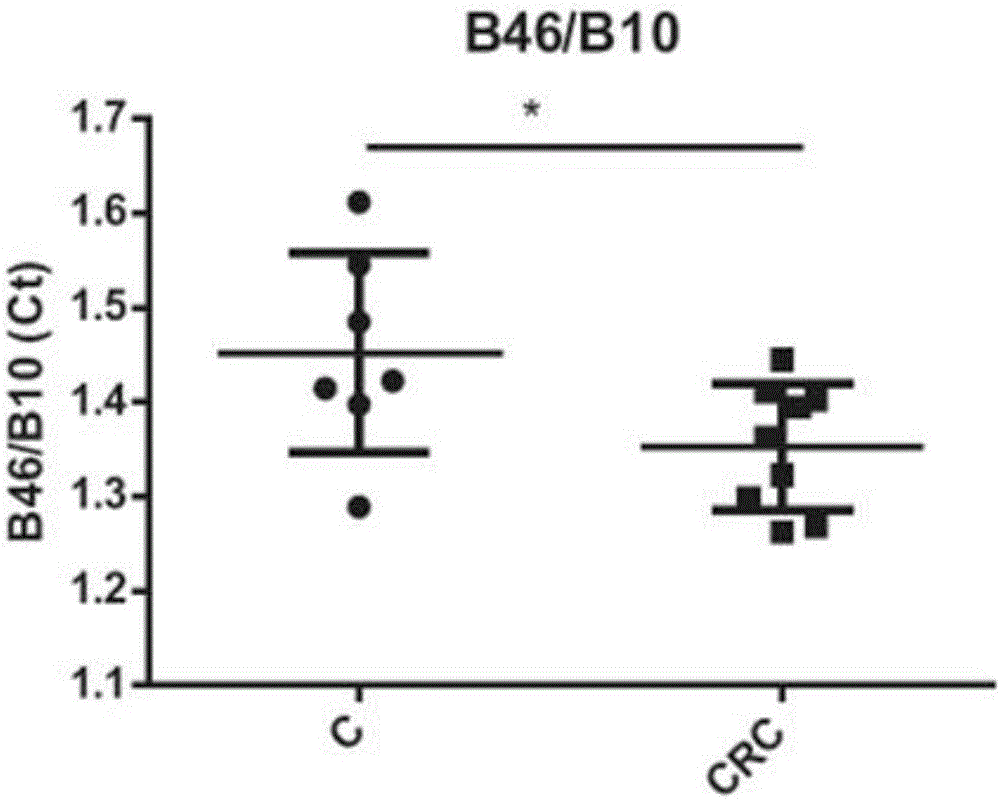
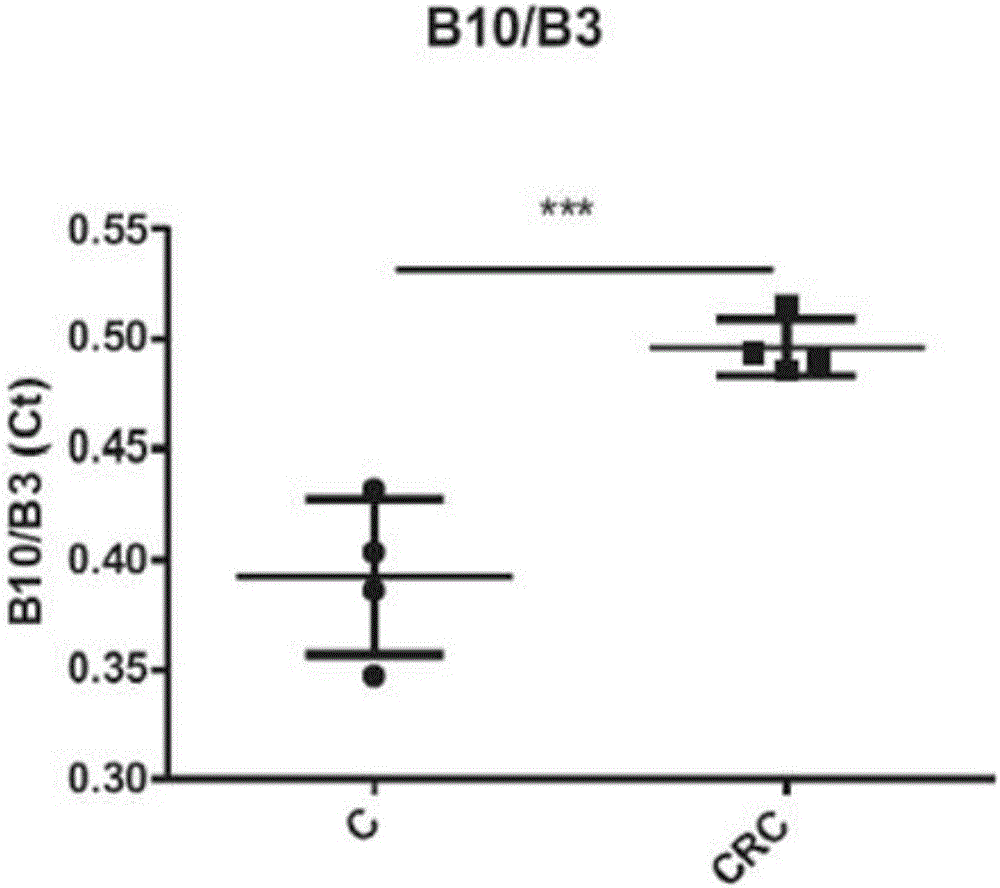
[0222] Example 3: Analysis of 16S rDNA biomarkers in colorectal cancer patients VS healthy subjects
[0223] A total of 16 stool samples from 7 controls and 9 patients with colorectal cancer were analyzed. The quantitative analysis of each bacterial marker as described in Example 1 in the analysis of stool samples was expressed as Ct value. Ct (cycle threshold) was defined as the number of q-PCR cycles required for the fluorescence signal to exceed the threshold. The Ct level is inversely proportional to the logarithm of the target nucleic acid concentration in the sample (ie, the lower the CT level, the higher the amount of target nucleic acid in the sample). Real-time analysis was performed over 40 amplification cycles. The results obtained are shown in Tables 1 and 2 below. Table 1 is the absolute value of Ct, and Table 2 is the ratio of Ct. A two-sample t-test was used.
[0224] Table 1: Absolute value of Ct
[0225]
[0226]
[0227] *Determination of accuracy...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com