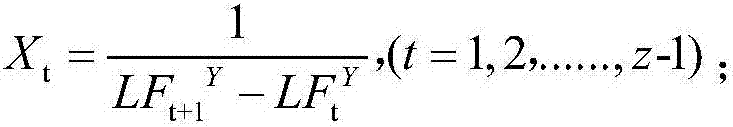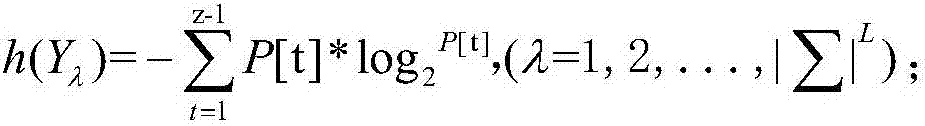DNA sequence cluster of Locality Sensitive Hashing based on standard entropy
A locally sensitive hashing and DNA sequence technology, applied in computer components, instruments, calculations, etc., can solve problems such as unstable clustering results, high time complexity, and difficult calculation of sequence data
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
[0029] Such as figure 1 Shown, the present invention is based on the DNA sequence clustering of the standard entropy local sensitive hash, and it comprises the following steps:
[0030] (1) The entire sequence to be tested is sequenced by second-generation sequencing technology to obtain a batch of short DNA fragments, and each short fragment is called a DNA fragment sequence;
[0031] (2) The letter set in the DNA fragment sequence is {A, C, G, T}, |∑| represents the number of letters in the letter set, initialize the word length L of the word to be processed, and use a fixed The length of the sliding window to obtain the set of words Y to be processed, the number of words Y to be processed in the set of words Y to be processed is |∑| L , according to the position information X of each word to be processed t Calculate its entropy value h;
[0032] The position information X of the word to be processed t Refers to the reciprocal of the distance between the two correspondin...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com