Method for inducing target protein degradation by bifunctional molecules
A technology for targeting ligands and compounds, which is applied in or administered to animals or humans to induce induction
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
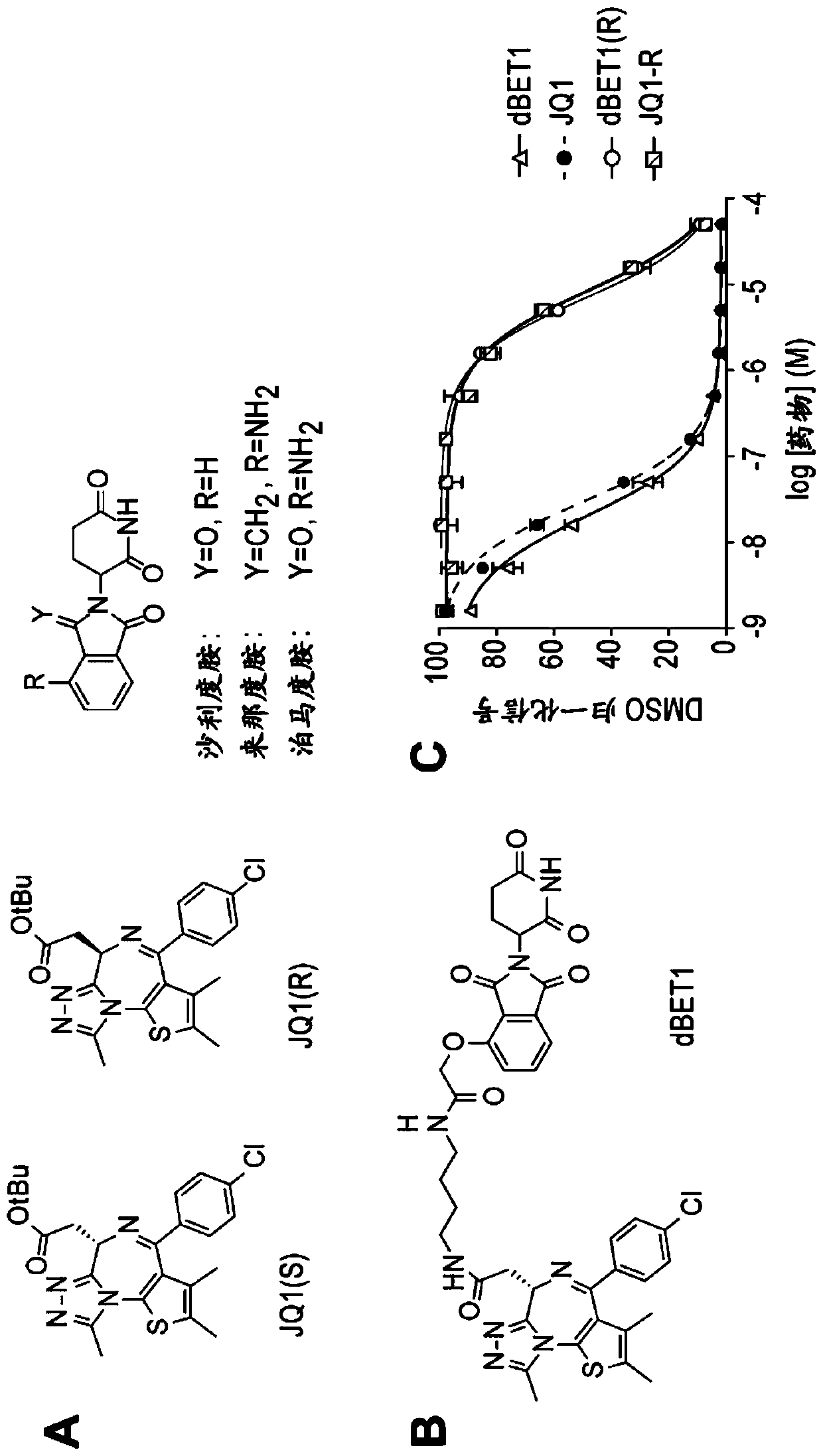
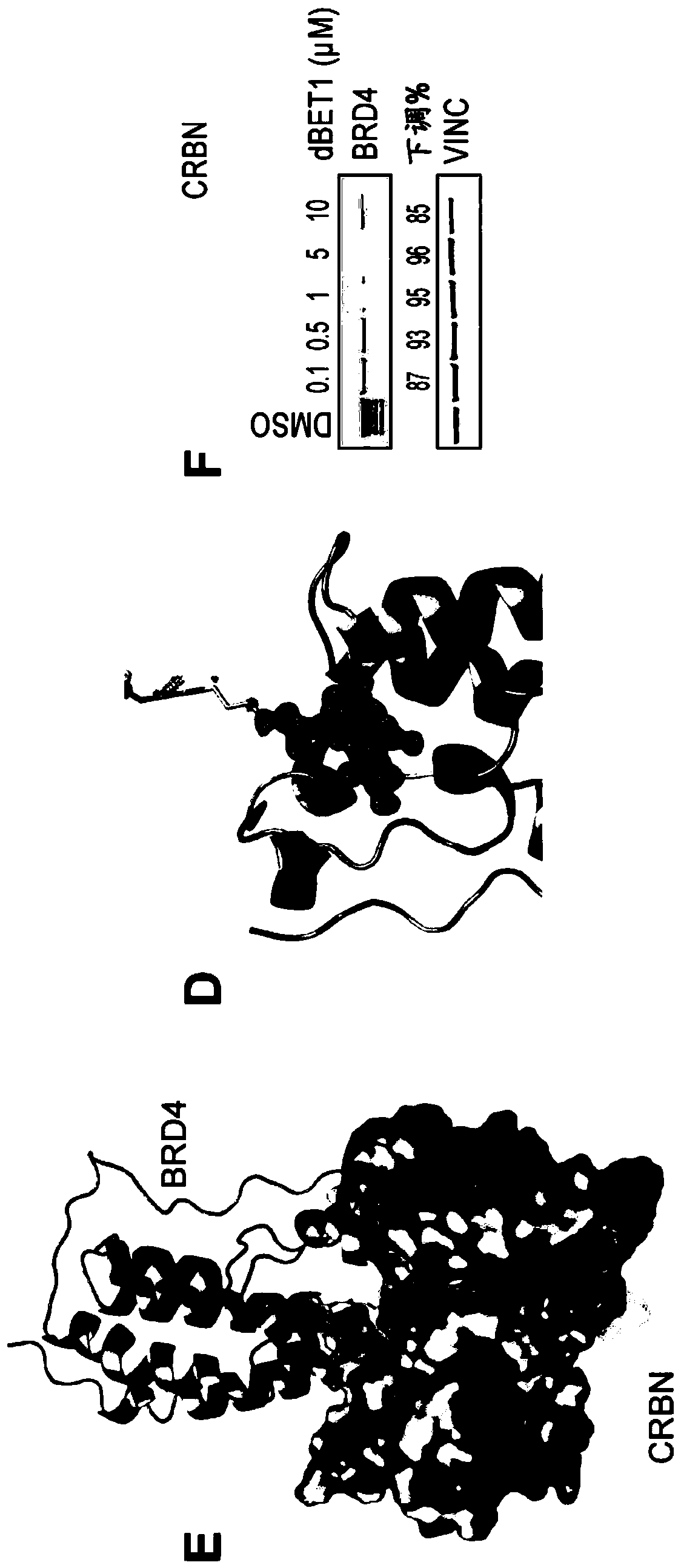
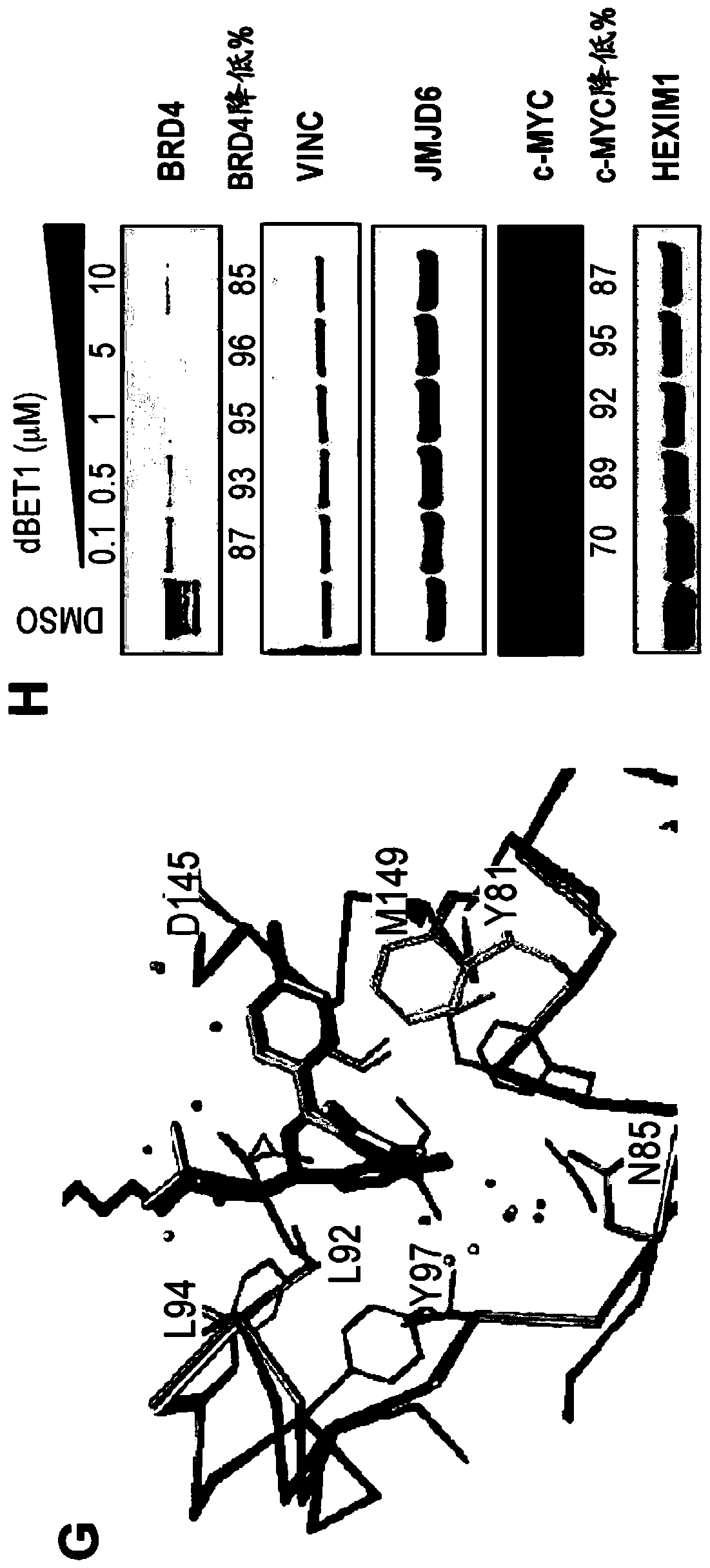
[1059] Example 1: Synthesis of dBET1
[1060]
[1061] (1) Synthesis of JQ-acid
[1062] JQ1 (1.0 g, 2.19 mmol, 1 equivalent) was dissolved in formic acid (11 mL, 0.2 M) at room temperature and stirred for 75 hours. The mixture was concentrated under reduced pressure to obtain a yellow solid (0.99 g, quantitative yield), which was used without purification. 1 H NMR (400MHz, methanol-d 4 )δ7.50–7.36(m,4H),4.59(t,J=7.1Hz,1H),3.51(d,J=7.1Hz,2H), 2.70(s,3H), 2.45(s,3H), 1.71 (s, 3H). LCMS 401.33 (M+H).
[1063] Synthesis of N-(4-aminobutyl)-2-((2-(2,6-dioxopiperidin-3-yl)-1,3-dioxoisoindoline- according to the previously published method 4-yl)oxy)acetamide trifluoroacetate (Fischer et al. Nature 2014, 512, 49).
[1064] (2) Synthesis of dBET1
[1065] Combine JQ-acid (11.3mg, 0.0281mmol, 1 equivalent) and N-(4-aminobutyl)-2-((2-(2,6-dioxopiperidin-3-yl)-1,3 -Dioxoisoindolin-4-yl)oxy)acetamide trifluoroacetate (14.5mg, 0.0281mmol, 1 equivalent) was dissolved in DMF (0.28mL, 0.1M) at room...
Embodiment 2
[1066] Example 2: Synthesis of dBET4
[1067]
[1068] Add N-(4-aminobutyl)-2-((2-(2,6-dioxopiperidin-3-yl)-1,3-dioxoisoindoline-4- (R)-JQ-acid (in a method similar to JQ-acid) was added to a 0.1M solution of acetamide trifluoroacetate in DMF (0.438mL, 0.0438mmol, 1.2 equivalents) R)-JQ1 preparation) (14.63mg, 0.0365mmol, 1 equivalent). DIPEA (19.1 microliters, 0.1095 mmol, 3 equivalents) and HATU (15.3 mg, 0.0402 mmol, 1.1 equivalents) were added, the mixture was stirred for 24 hours, then diluted with MeOH and concentrated under reduced pressure. The crude material was purified by preparative HPLC to obtain a yellow solid (20.64 mg, 0.0263 mmol, 72%). 1 H NMR (400MHz, methanol-d 4 )δ7.79(dd,J=8.4,7.4Hz,1H),7.51(d,J=7.3Hz,1H),7.47–7.39(m,5H),5.11–5.06(m,1H),4.75(s ,2H), 4.68(dd,J=8.8,5.5Hz,1H), 3.47–3.31(m,5H), 2.83–2.65(m,7H), 2.44(s,3H), 2.13–2.06(m,1H ), 1.68(s, 3H), 1.67-1.60(m, 4H). 13 C NMR(100MHz,cd 3 od)δ174.43,172.40,171.29,169.92,168.24,167.82,166.71,156.31,153.14,1...
Embodiment 3
[1069] Example 3: Synthesis of dBET3
[1070]
[1071] Add N-(2-aminoethyl)-2-((2-(2,6-dioxopiperidin-3-yl)-1,3-dioxoisoindoline-4- A 0.1 M solution of oxy)acetamide trifluoroacetate in DMF (0.475 mL, 0.0475 mmol, 1.2 equivalents) was added to JQ-acid (15.86 mg, 0.0396 mmol, 1 equivalent). Then DIPEA (20.7 microliters, 0.1188 mmol, 3 equivalents) and HATU (16.5 mg, 0.0435 mmol, 1.1 equivalents) were added, the mixture was stirred for 24 hours, and then purified by preparative HPLC to obtain a yellow solid (22.14 mg, 0.0292 mmol, 74%). 1 H NMR (400MHz, methanol-d 4 )δ7.82–7.75(m,1H),7.52–7.32(m,6H),5.04(dd,J=11.6,5.5Hz,1H), 4.76(d,J=3.2Hz,2H), 4.66(d ,J=6.6Hz,1H),3.58–3.35(m,6H), 2.78–2.58(m,6H), 2.48–2.41(m,3H), 2.11–2.02(m,1H), 1.70(d,J = 11.8Hz, 3H). 13 C NMR(100MHz,cd 3 od)δ174.38,171.26,171.19,170.26,168.86,168.21,167.76,166.72,156.27,153.14,138.44,138.36,138.19,134.87,133.71,132.31,131.57,131.51,129.90,129.86,121.81,119.36,117.95, 54.83, 50.52, 40.09, 39.76, 38.30, 32.09,...
PUM
| Property | Measurement | Unit |
|---|---|---|
| diameter | aaaaa | aaaaa |
| diameter | aaaaa | aaaaa |
| diameter | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com