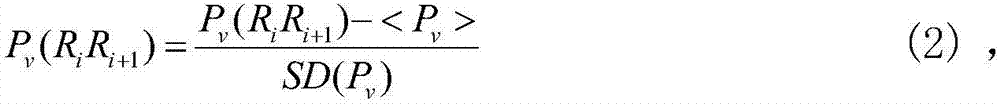Nucleosome classification forecasting method based on convolutional neural network
A technology of convolutional neural network and classification prediction, which is applied in the field of classification prediction of genetics and can solve problems such as the limitation of positioning accuracy
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
[0046] The content of the present invention will be further described below in conjunction with the accompanying drawings and embodiments, but the present invention is not limited.
[0047] Example:
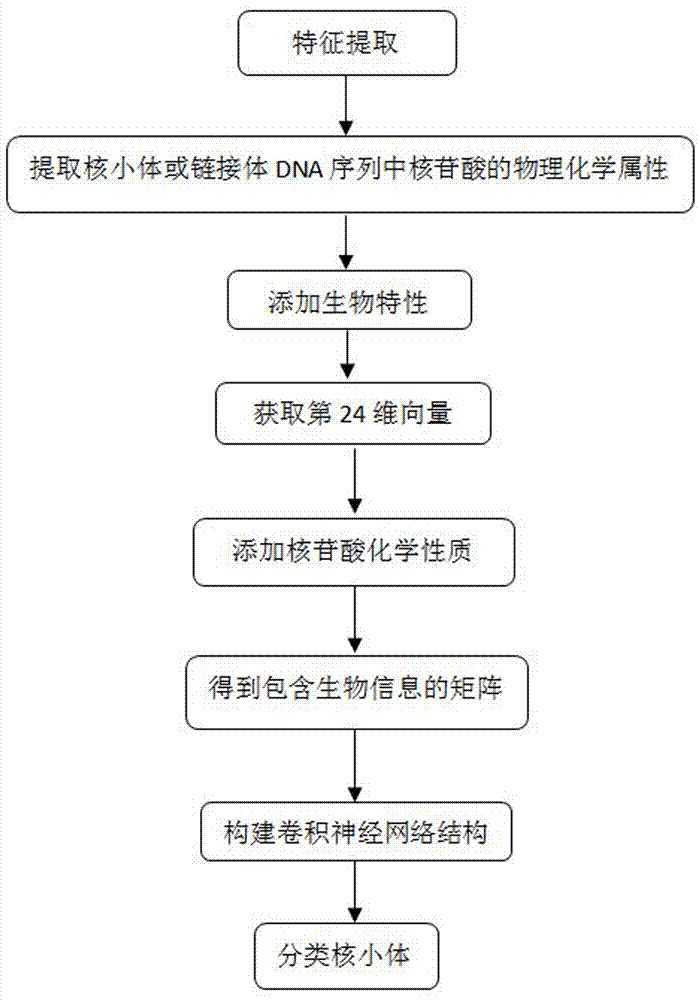
[0048] refer to figure 1 , a method for predicting nucleosome classification based on a convolutional neural network, comprising the following steps:
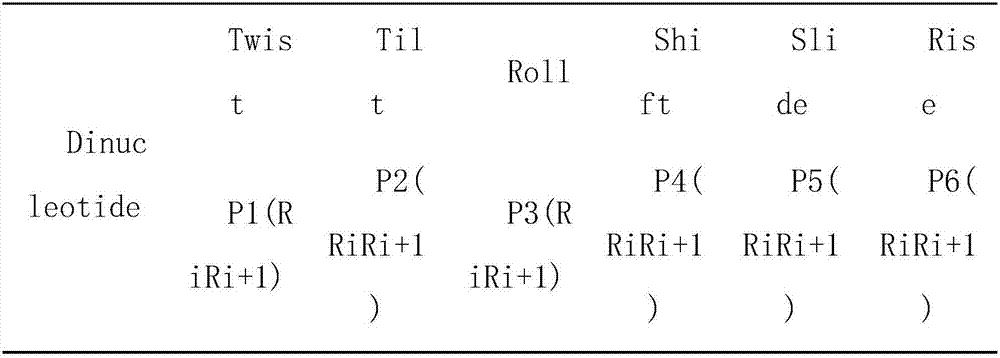
[0049] 1) Feature extraction: select the DNA sequences of the nucleosomes or linkers of Homo sapiens, nematodes and Drosophila melanogaster in the UCSC genome database. Refers to the base pair, which converts the 16 combinations of the dinucleotide ATCG in the DNA sequence of each nucleosome or linker into a 16-dimensional vector through one-hot encoding. The feature vector is expressed as formula (1) :
[0050] x i =(P i,1 ,P i,2 ,...,P i,16 ) T (1)
[0051] x i Indicates the eigenvector of the i-th nucleosome or link body at this time, P i,1 ,P i,2 ,...,P i,16 Represents one-hot encoding of 16 combinations of dinuc...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com