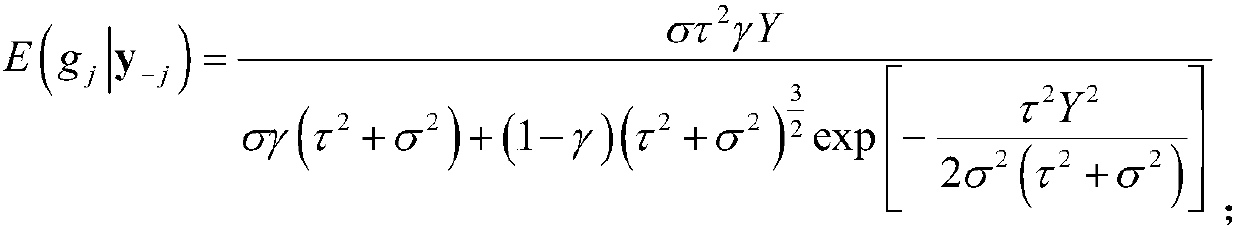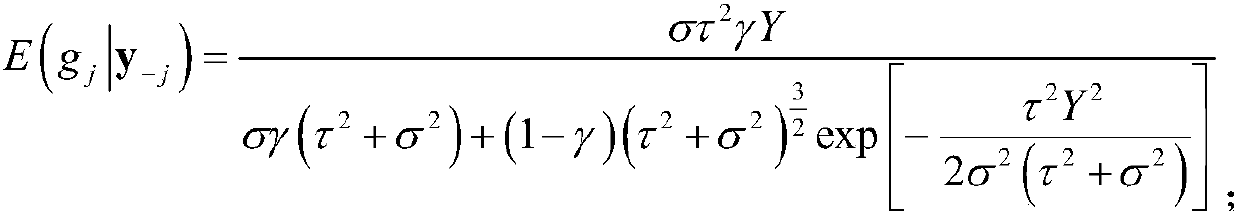New fast Bayes method estimating genomic estimated breeding value
A genome and breeding value technology, applied in computing, instrumentation, electrical and digital data processing, etc., can solve problems such as numerical out-of-bounds, inability to guarantee the convergence of SNP effect values, etc. Effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
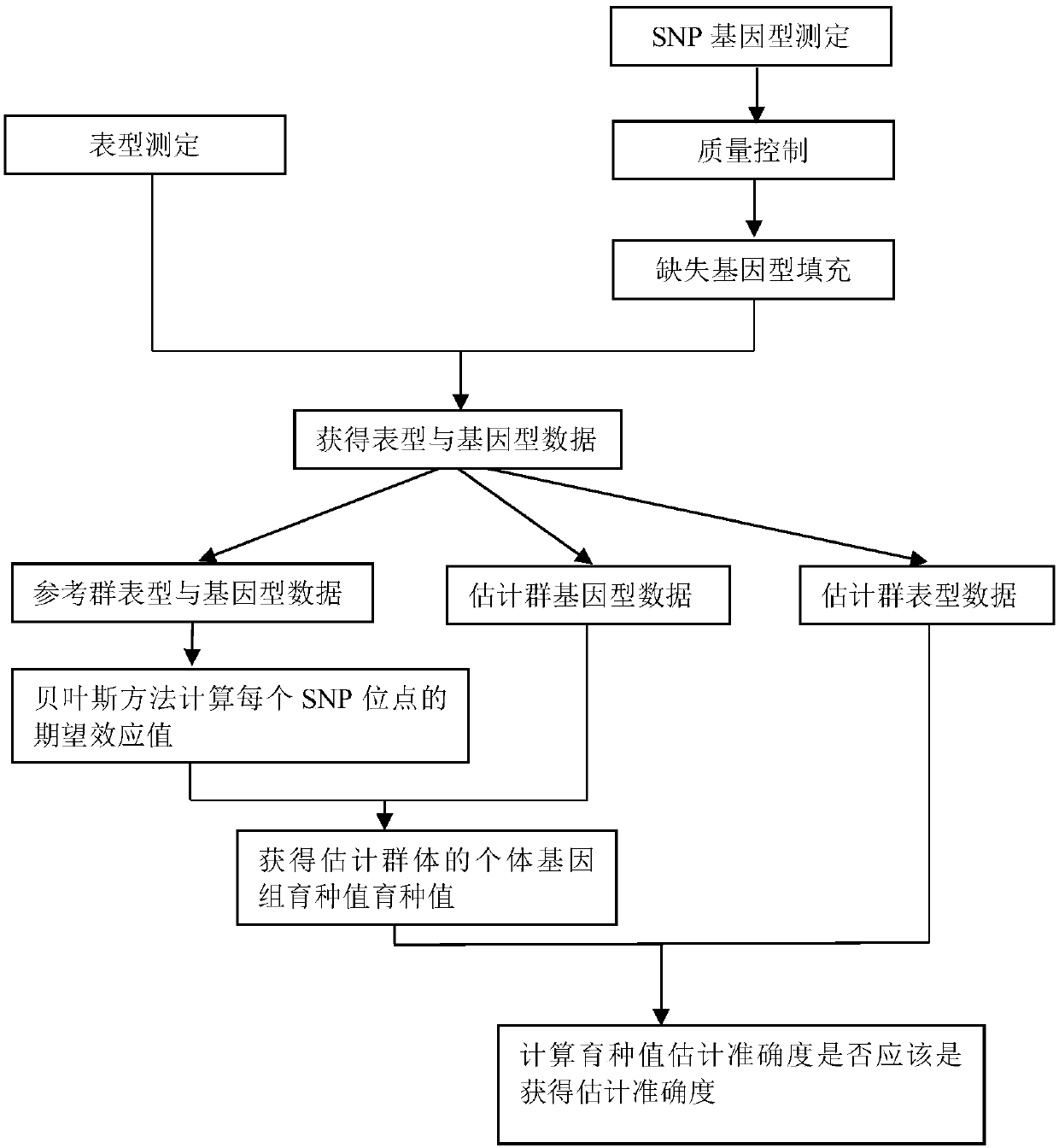
[0094] The test data is large yellow croaker, 30 male fish and 30 female fish are kept in the same tank, and the artificial induction technology is used to make all parent fish release sperm or eggs at almost the same time, so the offspring have the same age. When the offspring grew to 2 years old, 500 offspring (including 237 males and 263 females) were randomly selected as the experimental materials of this study, and the phenotypic data of the two traits (drained weight and net weight rate) were determined .
[0095] GBS (genotyping-by-sequencing) technology was used to sequence the genomes of all individuals to be studied, and the qualified SNP sites were screened. After screening MAF>0.05, Hardy-Weinberg balance test P-value>0.001, site deletion rate Below 20% of the SNP markers, a total of 29748 qualified SNP sites in the genome were obtained. For missing sites, the imputation program of software Beagle version 3.3.2 was used to fill in.
[0096] All 500 individuals we...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com