Brand new distributed and privatized miRNA-disease association prediction method
A forecasting method and distributed technology, applied in the fields of molecular biology and life sciences, can solve problems such as unsatisfactory forecasting accuracy
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
[0068] The present invention will be described in detail below in combination with specific embodiments.
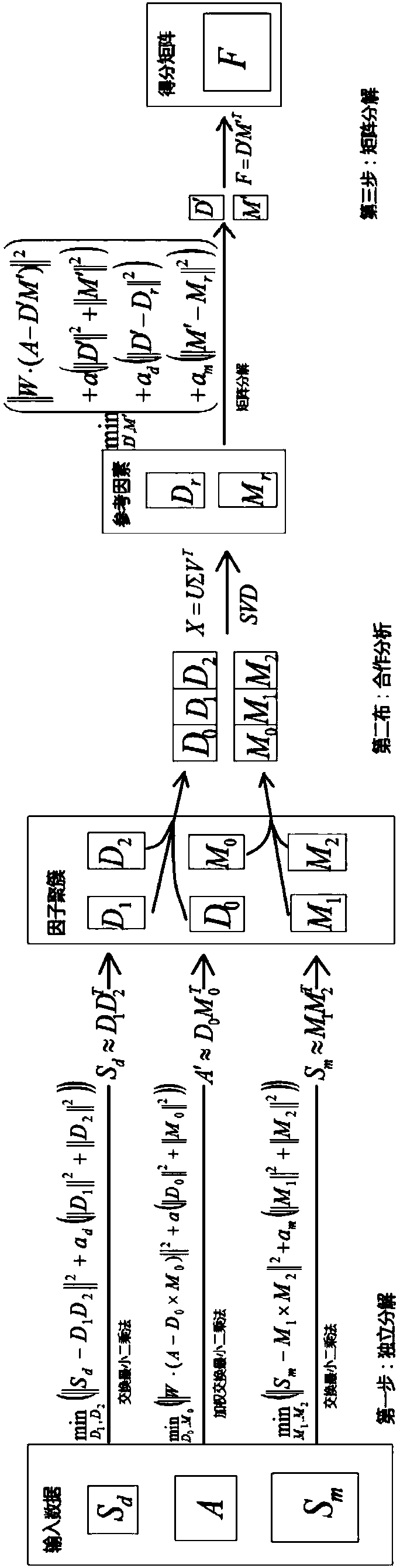
[0069] Such as Figure 1-3 As shown, the present invention is a brand-new distributed and privatized miRNA-disease association prediction framework, which is specifically implemented according to the following steps.
[0070] Step 1. Data input.
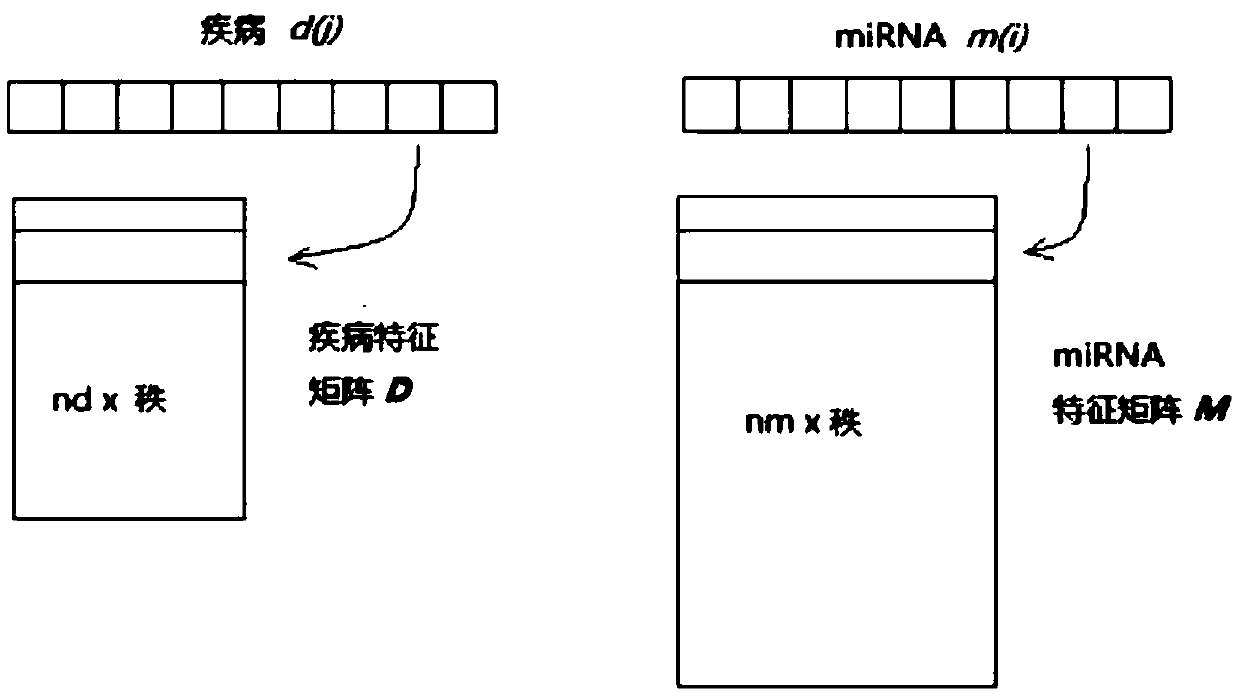
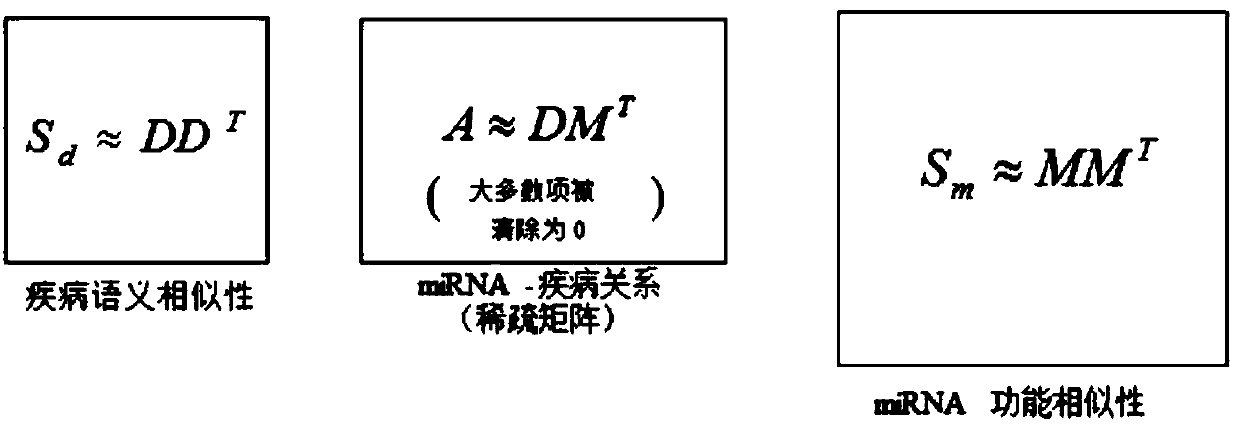
[0071] First, in order to obtain the association between miRNAs and diseases, three matrices are used as input, representing miRNA-disease association, miRNA functional similarity, and disease semantic similarity, respectively.
[0072] The HMDD database was used as a source of human miRNA-disease relationships, with 5430 experimentally validated records, partially revealing the links between 383 diseases and 495 miRNAs in the latest version. An association matrix A was constructed to describe miRNA-disease associations. A(d(i); m(j))=1 means that miRNAm(j) is experimentally confirmed to be associated with disease d(i), other...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com