Preparation method and application of Cre and Flp dependent retrograde tracing recombinant pseudo-rabies virus
A technology of pseudorabies virus and recombinant virus, which is applied in the biological field and can solve the problems of lack of recombinant pseudorabies virus, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
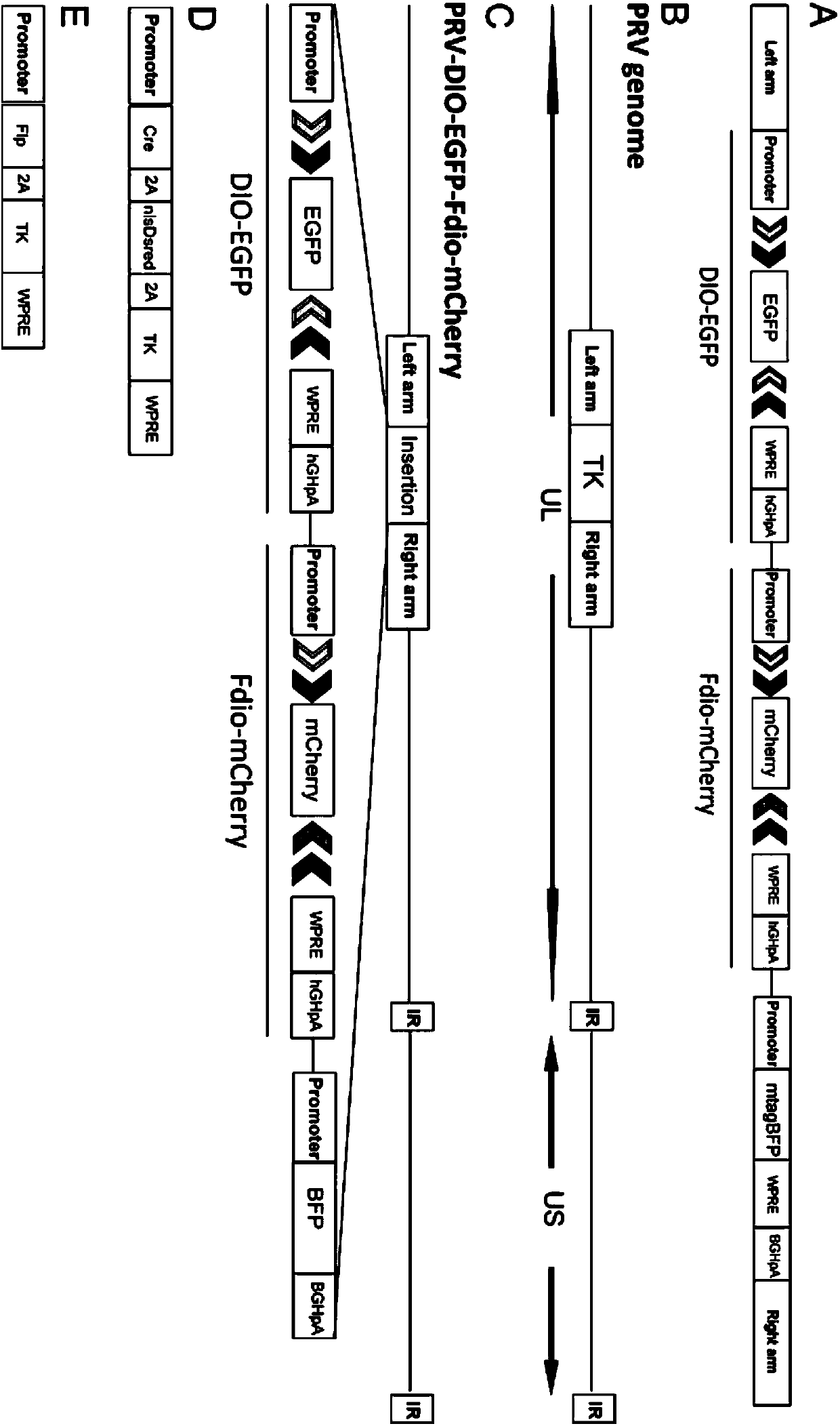
[0042] Embodiment 1: a kind of Cre and Flp system-dependent reverse tracing recombinant pseudorabies virus preparation method, comprising the following steps:
[0043] 1.1 Cre and Flp-dependent clones expressing green fluorescent protein and red fluorescent protein and containing homology arms:
[0044] 1.1.1 Clones capable of Cre-dependent expression of green fluorescent protein
[0045] The PCR method was used to amplify EGFP (see SEQ ID NO.1 for the sequence) and Ubc promoter (see SEQ ID NO.2 for the sequence). The primers for amplifying the corresponding sequence are as follows respectively: primers for the DNA fragment EGFP: SEQ ID NO:3 and SEQ ID NO:4, the template is pEGFP; primers for the DNA fragment Ubc promoter: SEQ ID NO:5 and SEQ ID NO:6 , the template is FUGW(14883#, addgene). All PCR primers used in the present invention were synthesized by Sangon Bioengineering (Shanghai) Co., Ltd. First use AscI and NcoI to double cut pAAV-EF1a-double floxed-hChR2(H134R)-EY...
Embodiment 2
[0063] Example 2: A Cre and Flp system-dependent reverse tracer Recombinant pseudorabies virus expresses green fluorescent protein and red fluorescent protein depending on Cre and Flp respectively:
[0064] At present, there is no reverse visualization system that has the ability to express both Cre and Flp-dependent green fluorescent protein and red fluorescent protein in a recombinant pseudorabies virus at the same time. Fluorescent proteins and DsRed capable of analyzing:
[0065] On the one hand, by transfecting pNrl-Cre (can express Cre, 13780#, addgene) to BHK21 cells, take 5 μ l of the recombinant pseudorabies virus PRV527 prepared in Example 1 after 24 hours (the virus titer is 1.5×10 7 PFU / ml) to infect BHK21 cells expressing Cre, 37°C, 5% (v / v) CO 2 They were cultured in an incubator, and the expression of fluorescence was observed using the same exposure parameters after infection.
[0066] Experimental results:
[0067] figure 2 Middle A: Cells expressing Cre ...
Embodiment 3
[0073] Embodiment 3: A Cre and Flp system-dependent reverse tracer recombinant pseudorabies virus stability analysis:
[0074] In order to analyze the stability of PRV527 carrying Cre and Flp-dependent elements, take 5 μl of PRV527 prepared in Example 1 (the virus titer is 1.5×10 7 PFU / ml) (as P0) to infect BHK21 cells, collect the supernatant (as P1) after 2 days of infection, carry out passage on BHK21 cells for 10 generations according to the above method, collect the virus fluid of each generation, and perform plaqueassay to detect the morphology of plaque on the one hand and uniformity, on the other hand, the stability of EGFP in the process of virus passage and the ability of the virus to express fluorescence after infecting BHK21 cells in vitro were analyzed. The result is as image 3 As shown, the recombinant pseudorabies virus PRV527 was passed on BHK21 cells for 10 generations. Get the recombinant pseudorabies virus PRV527 of P10 generation to detect according to t...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com