Systems and methods for analyzing nucleic acid
A nucleic acid and sequence technology, applied in the field of tumor-specific biomarkers, which can solve problems such as incorrect prognosis or ineffective treatment, sequencing errors, and patients missing treatment opportunities.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment
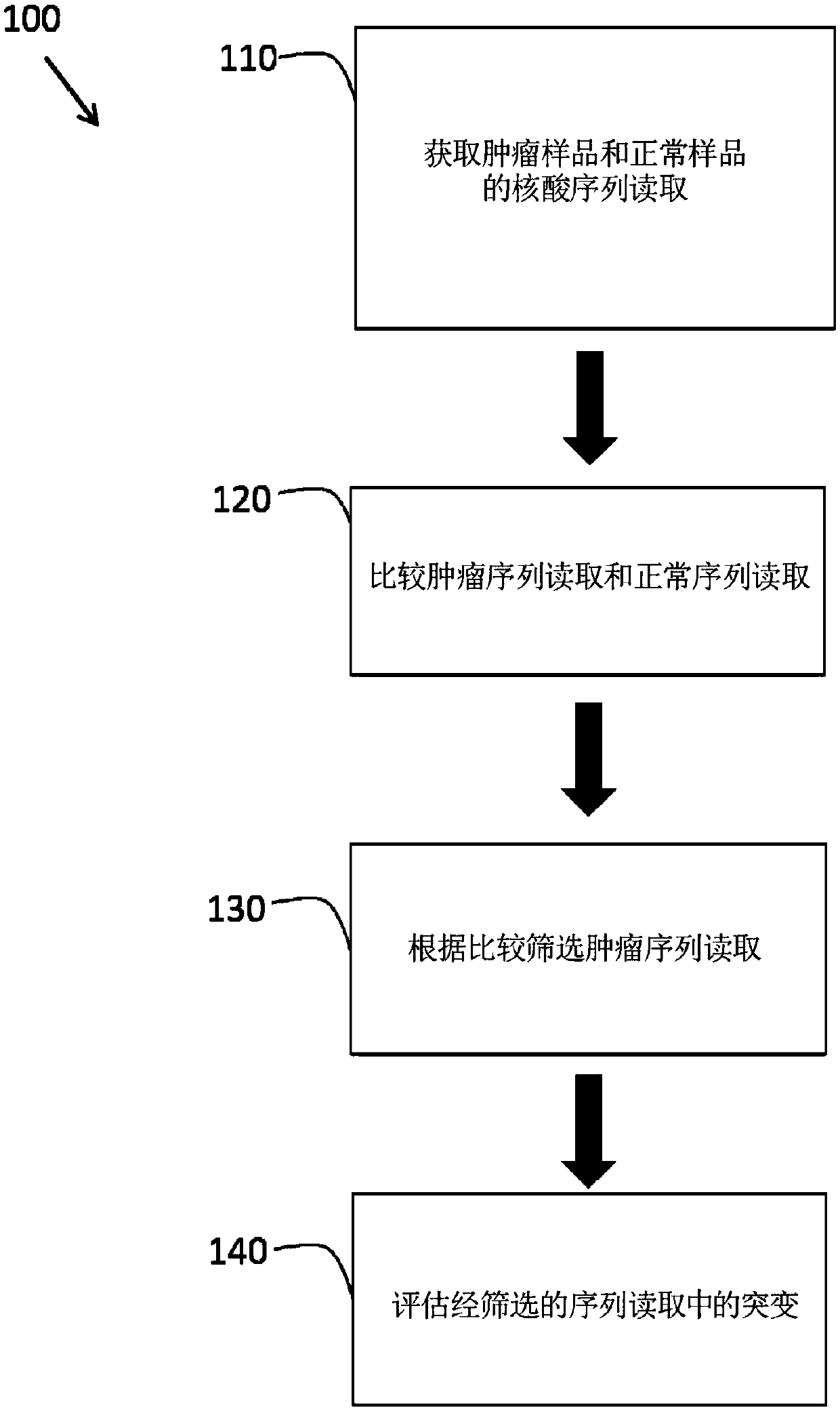
[0142] To assess the clinical utility of the large-scale cancer genome analysis disclosed herein, whole exome and targeted next-generation sequencing analyzes were performed in tumor and normal samples from cancer patients. Co- and separate analysis of matched tumor and normal data for somatic mutations, potential clinical actionability, and identification of altered susceptibility.
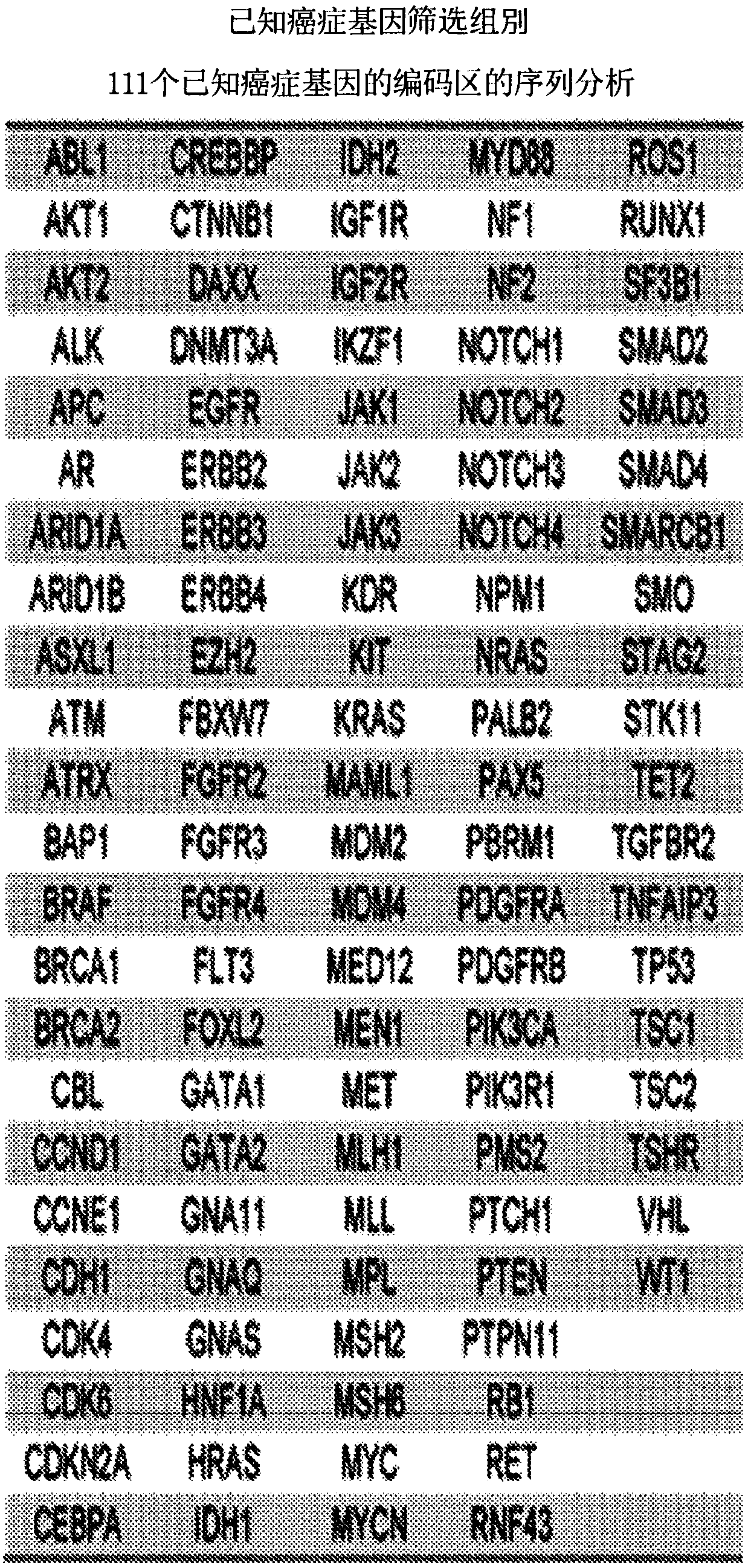
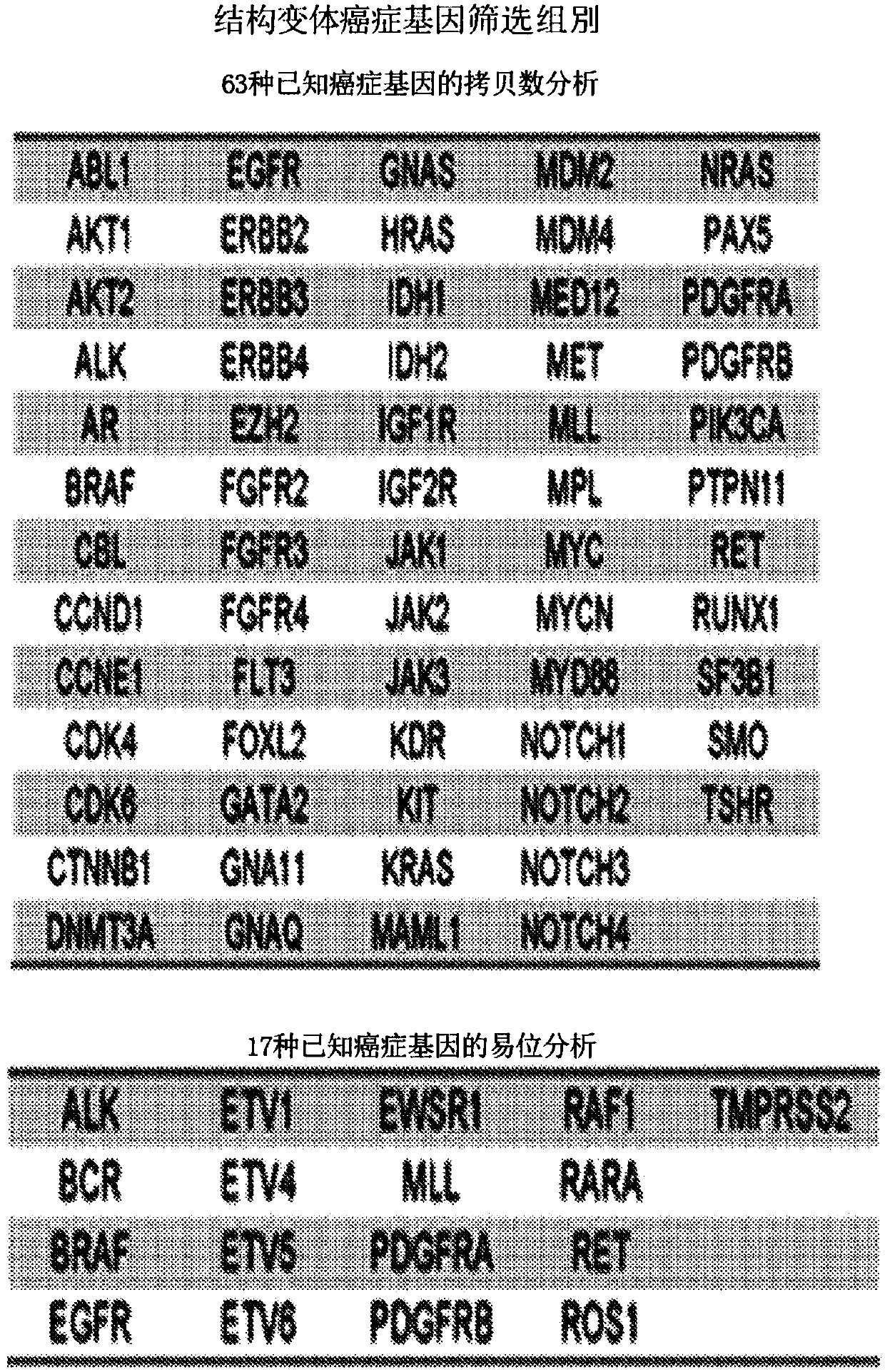
[0143] Eight hundred and fifteen (815) tumor-normal paired samples from patients with fifteen tumor types were comprehensively evaluated. Genome-wide or 111 targeted genes were identified using next-generation sequencing methods with >95% and >99% sensitivity and >99.9% specificity, respectively. These analyzes revealed an average of 140 and 4.3 somatic mutations per exome and on-target analysis, respectively. More than 75% of cases had somatic alterations in genes associated with known therapies or current clinical trials, and most actionable genes were infrequently altered in any tumor type. ...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Sensitivity | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com