Traceless gene editing method for trichoderma fungi
A gene and Trichoderma technology, which is applied in the field of Trichoderma without traces in the field of gene cycle editing, can solve the problems of complex internal mechanism and inability to explain the comprehensive results of interaction of multi-gene products.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
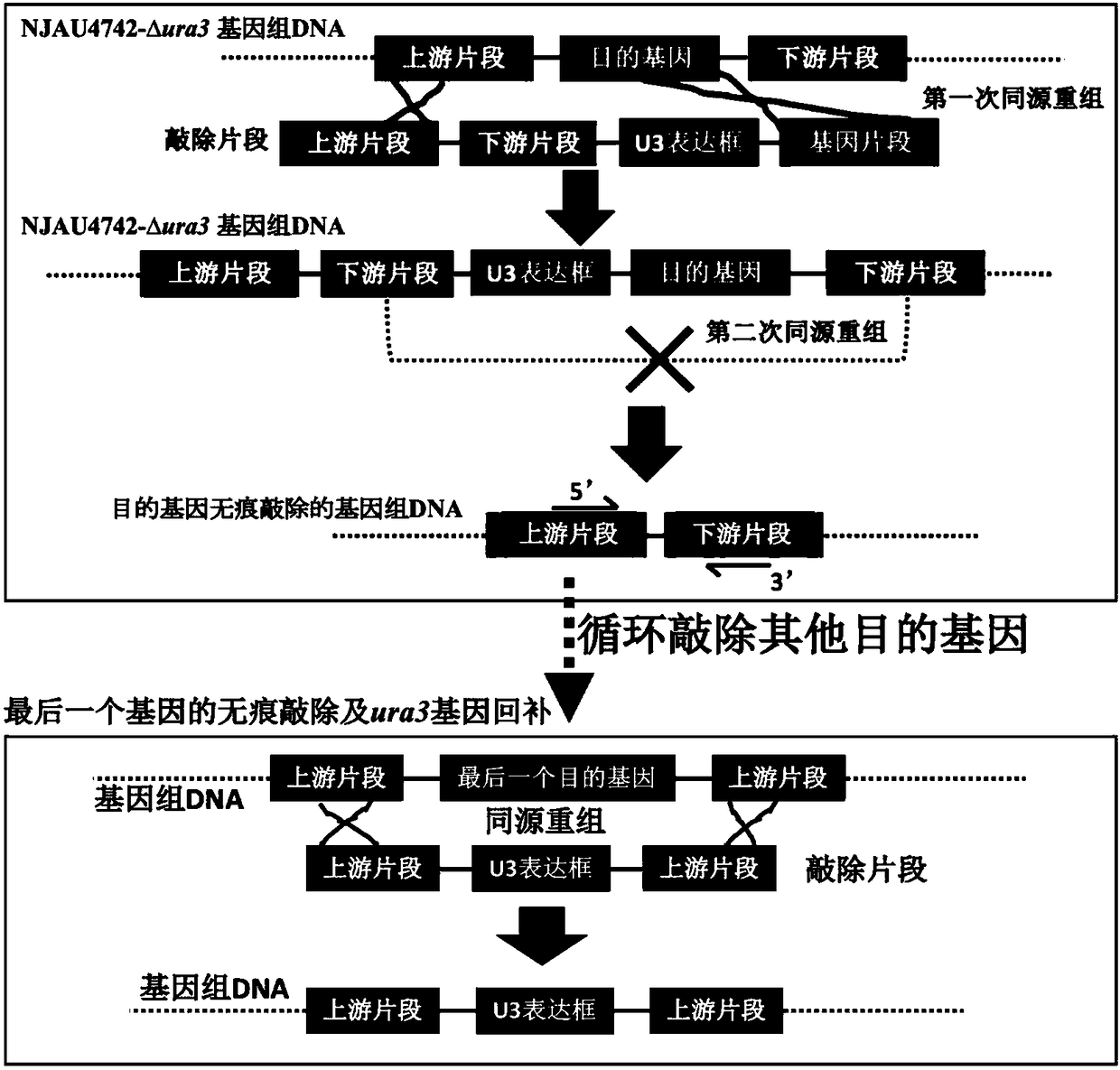
[0044] Implementation 1: Construction of auxotrophic strain NJAU4742-Δura3 traceless mutant of Trichoderma Guizhou NJAU4742 (1) Construction of knockout fragment
[0045] In the process of seamless knockout of ura3, the presence or absence of the gene itself is used as a screening marker for secondary homologous recombination, so the construction of the knockout fragment is slightly different. The knockout fragment consists of 4 different parts: upstream fragment + downstream fragment + HygB resistance gene expression box + gene fragment. Among them, the HygB resistance gene expression cassette can be transcribed and translated into hygromycin B phosphotransferase, making the original strain NJAU4742 resistant to hygromycin B. After the second homologous recombination, the HygB resistance gene expression cassette and ura3 Gene sequences will be deleted together, so 5-FOA can be used for reverse screening to obtain a traceless knockout mutant of the ura3 gene. In summary, in t...
Embodiment 2
[0053] Implementation 2: The traceless knockout of the xyr1 gene, which regulates lignocellulosic degrading enzymes in Trichoderma Guizhou NJAU4742
[0054] Trichoderma strain: Guizhou Trichoderma NJAU4742-Δura3 (NJAU4742-Δura3)
[0055] (1) Construction of knockout fragments
[0056] Knockout fragment amplification primers are: upstream fragment amplification primer x1_upF: GGCCTTGAAACGGTATGTCGA (SEQ ID NO.11) and x1_upR: CGTACACCACCATCACAGGGATATCAATAGGAGATGGCTGAACTGTGTG (SEQ ID NO.12); downstream fragment amplification primer x1_downF: CACACAGTTCAGCCATCTCCTATTGATATCCCCTGTGATGTQGTACG 和x1_downR:GCCATATTGATGTAAGGTAGCTCTCCTCACTTCCGCTTCACATAGACC(SEQ ID NO.14);U3表达框扩增引物U3box_F:GAGAGCTACCTTACATCAATATGGCGCGCAGATGTAGCGGTACATG(SEQ ID NO.15)和U3box_R:GGTACTATGGCTTAGATGGAATACCCCGTATTCTCTGCCCTTGTTGC(SEQ ID NO.16);基因片段扩增引物x1_geneF:GGGTATTCCATCTAAGCCATAGTACCCTTCTTCAGCCCTTGATCCACAC(SEQ ID NO. 17) and x1_geneR: CCTTGATTCACACGCAAATGTTCC (SEQ ID NO. 18). The final concentration of the knockout...
Embodiment 3
[0064] Implementation 3: Trichoderma Guizhou NJAU4742 carbon source inhibition regulatory gene cre1 traceless knockout and ura3 gene complement Trichoderma strain: Guizhou Trichoderma NJAU4742-Δura3-Δxyr1 (NJAU4742-Δura3-Δxyr1)
[0065] (1) Construction of homologous knockout fragments
[0066] The knockout fragment is: cre1 gene upstream fragment + U3 expression box + cre1 gene downstream fragment. The amplification primers are: upstream fragment amplification primer c1_upF: GGTGGGCAAAAAGGAACCTGG (SEQ ID NO.21) and c1_upR: GCCATATTGATGTAAGGTAGCTCTCAGAGATTATCCGCTGGTGGAGTG (SEQ ID NO.22); U3 expression box amplification primer U3box_F: GAGAGCTACCTTACATCAATATGGCGCGCAGATNOGTAGCGGTACATG (SEQ.2) U3box_R: GGTACTATGGCTTAGATGGAATACCCCGTATTCTCTGCCCTTGTTGC (SEQ ID NO. 24); downstream fragment amplification primer c1_downF: GGGTATTCCATCTAAGCCATAGTACCGCGCCTCGAATGACTTGATGAC (SEQ ID NO. 25) and c1_downR: GCCCTACGAGAATGTCGGTTC (SEQ ID NO. 26). The final concentration of the knockout fragmen...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com