Method for analyzing tumor mutation load on the basis of next-generation sequencing data of single sample
A mutation load and next-generation sequencing technology, which is applied in sequence analysis, electrical digital data processing, special data processing applications, etc., can solve the problems of increasing the cost of experiments and sequencing
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment
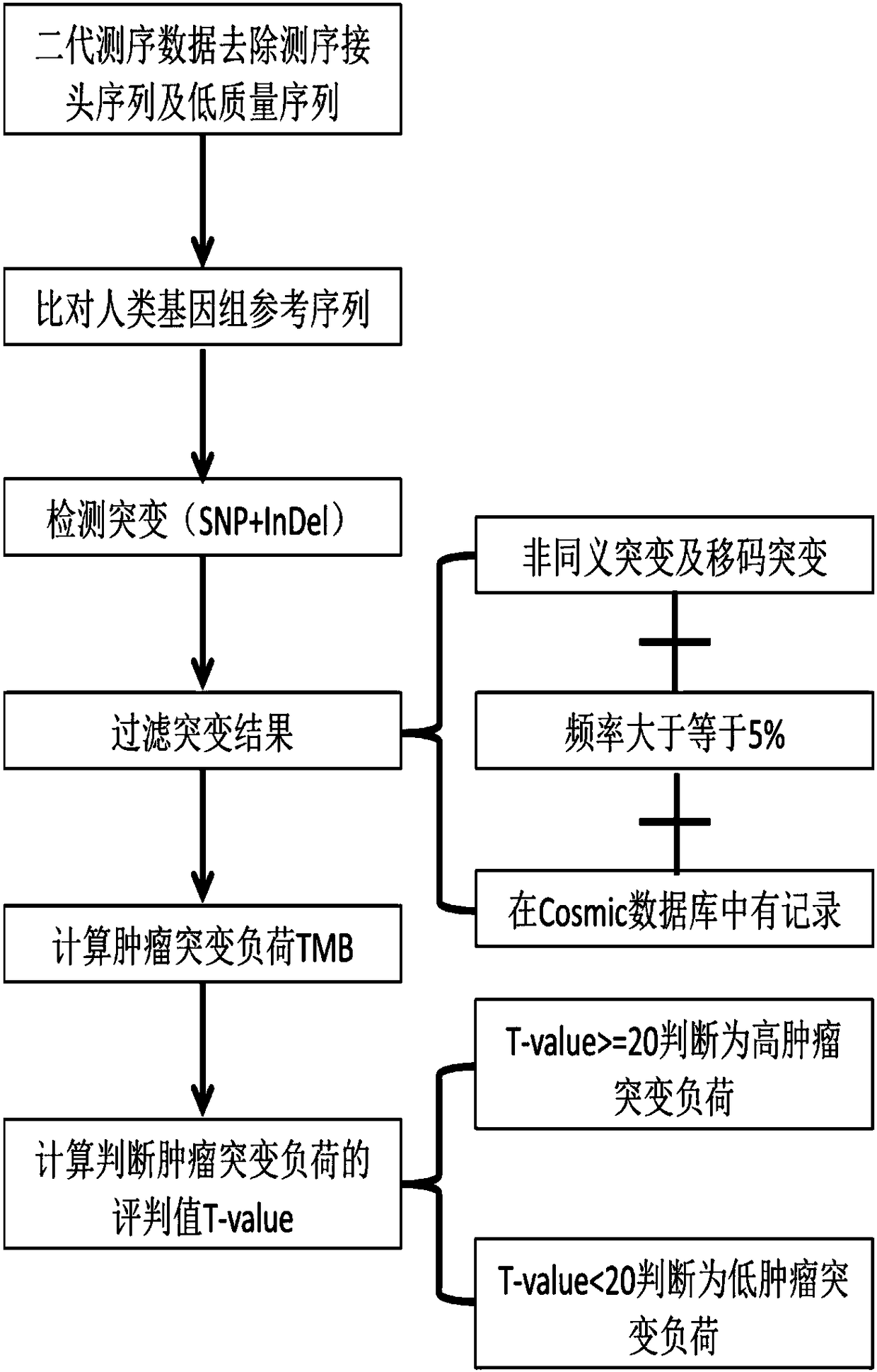
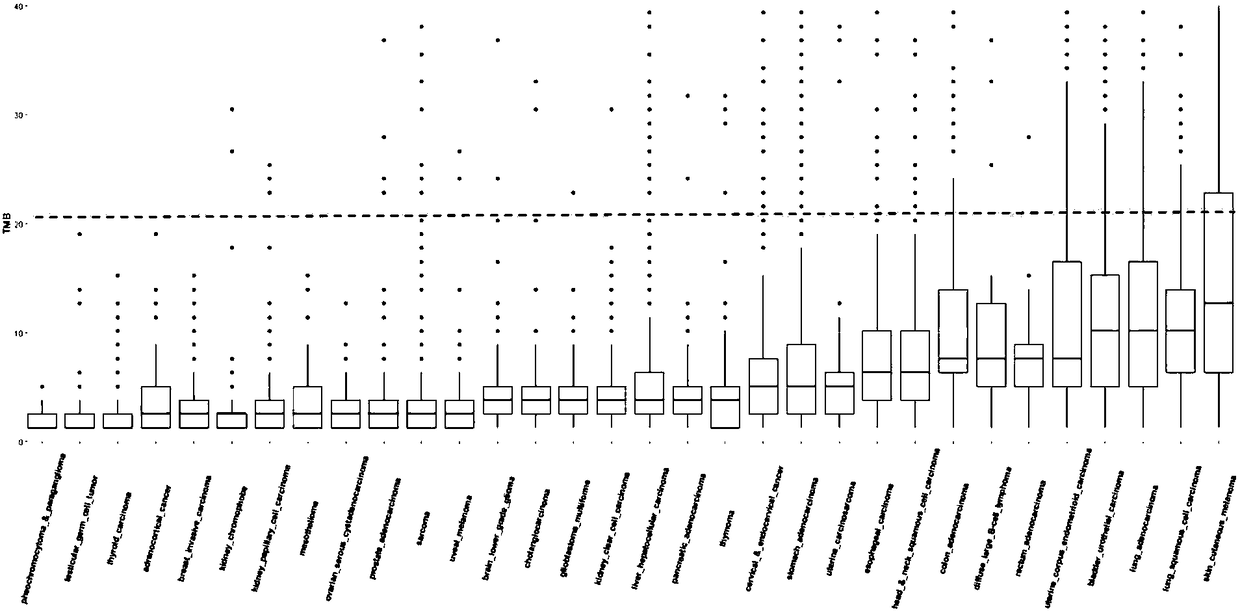
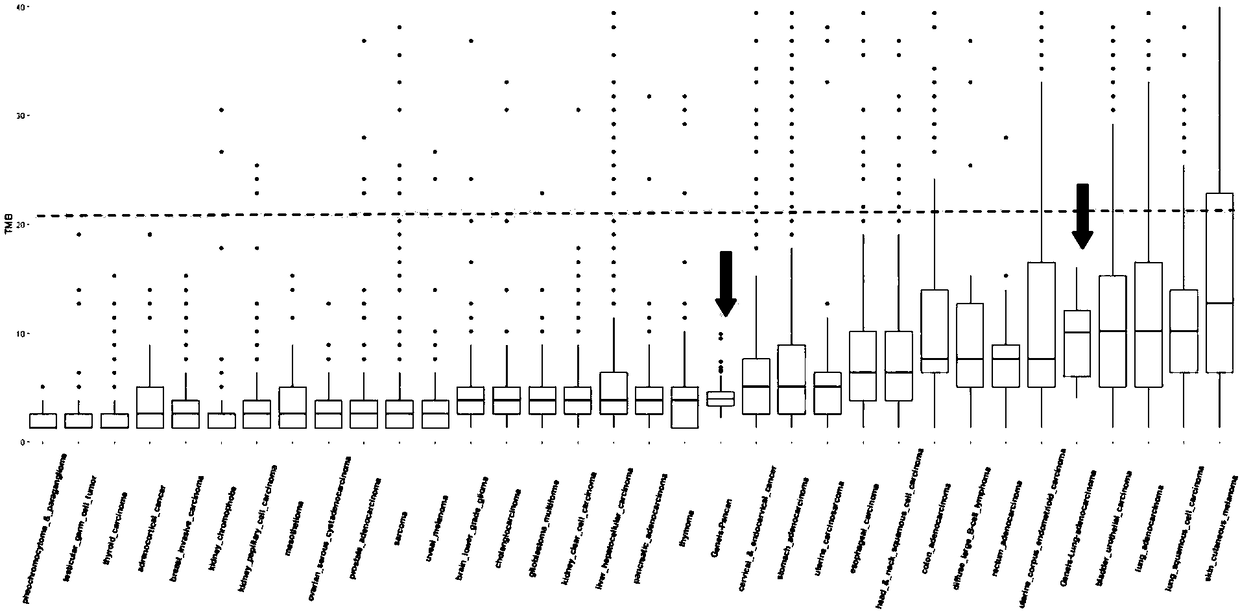
[0044] Select 75 pancreatic cancer tumor samples, use the Geneis-508 Gene Panel kit (the kit contains exon regions of 226 genes, with a total size of 787,296bp) for hybrid capture and use Illumina Nextseq for paired-end 151bp sequence read length Sequencing was performed to obtain the data set Data-A (data of 75 sequencing samples).
[0045] Select 33 cases of lung adenocarcinoma tumor samples, use the Geneis-38CF Gene Panel kit (the kit contains the exon regions of 8 genes, the total size is 49,861bp) for hybrid capture and use Illumina Nextseq for paired-end 151bp sequence reading Sequencing was carried out to obtain the data set Data-B (data of 33 sequencing samples).
[0046] Download the analysis results of somatic cell data from the TCGA (The Cancer Genome Atlas, https: / / cancergenome.nih.gov / ) database to obtain the data set Data-C.
[0047] The 226 gene exon regions of the Geneis-508 Gene Panel were used to screen the somatic mutation results of the data set Data-C and...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com