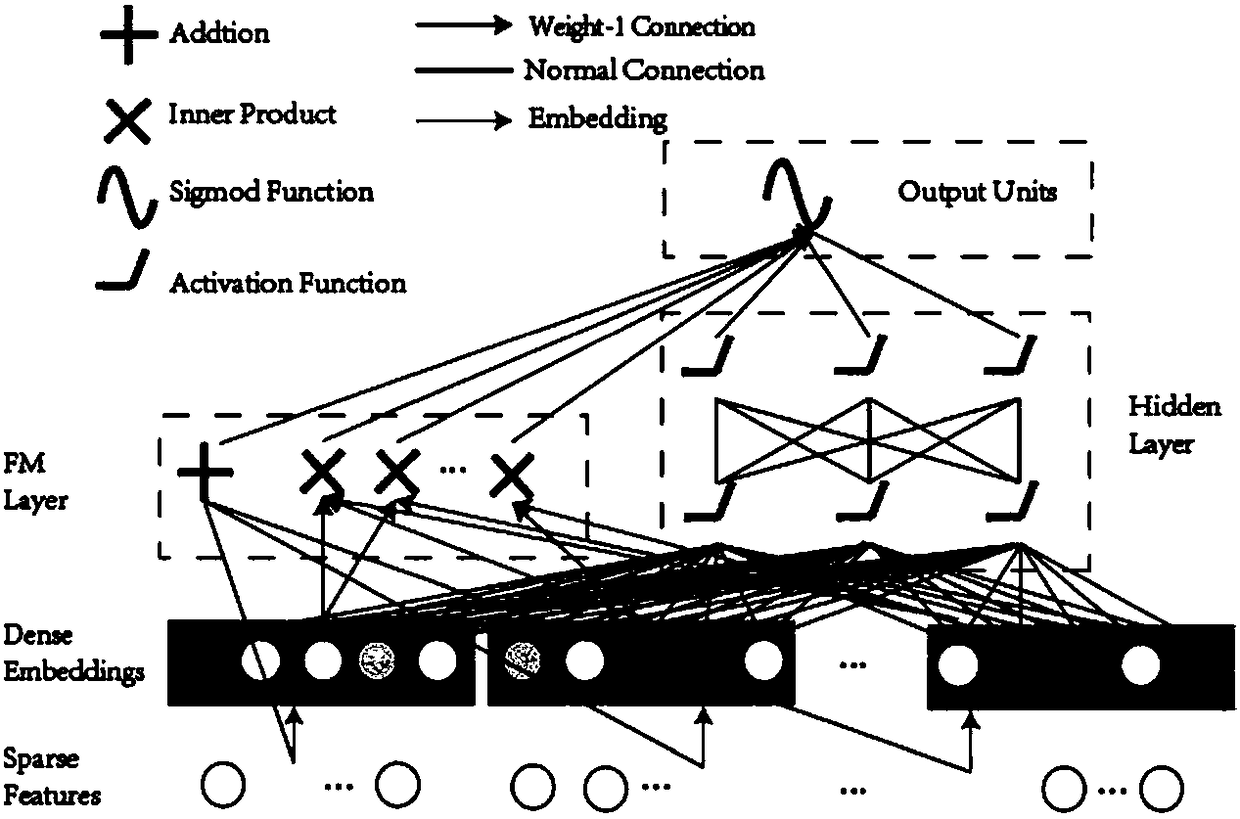
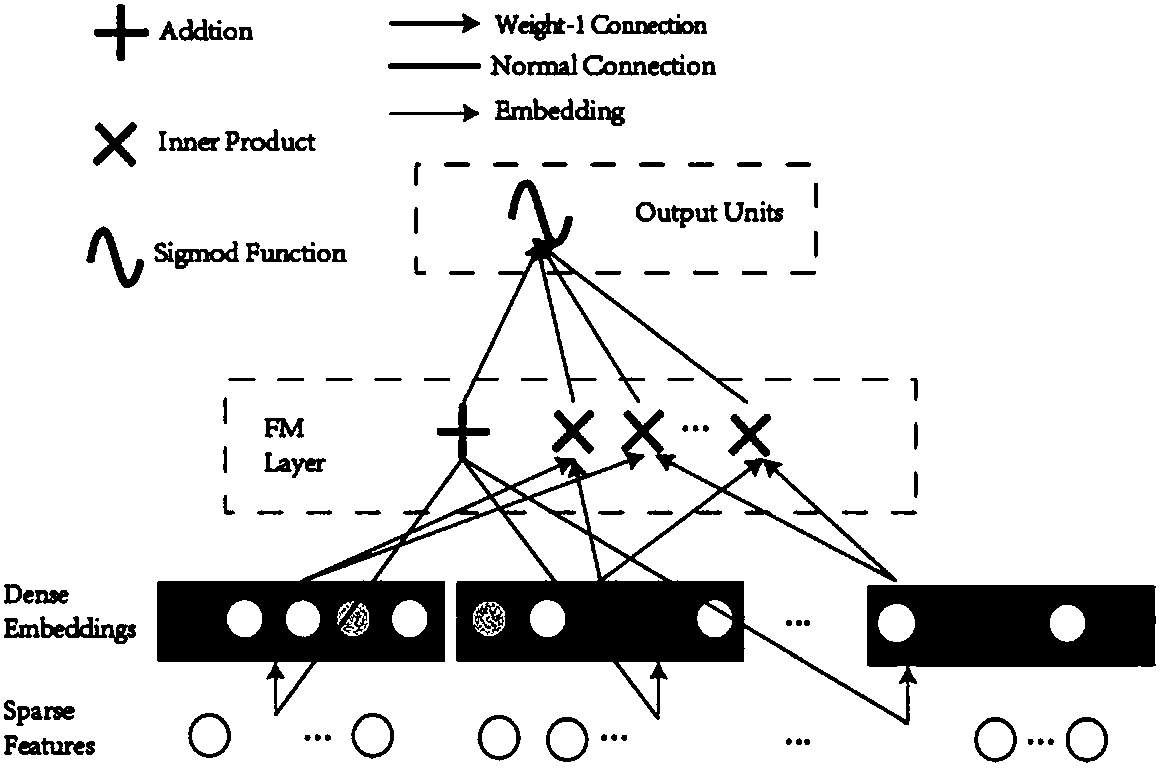
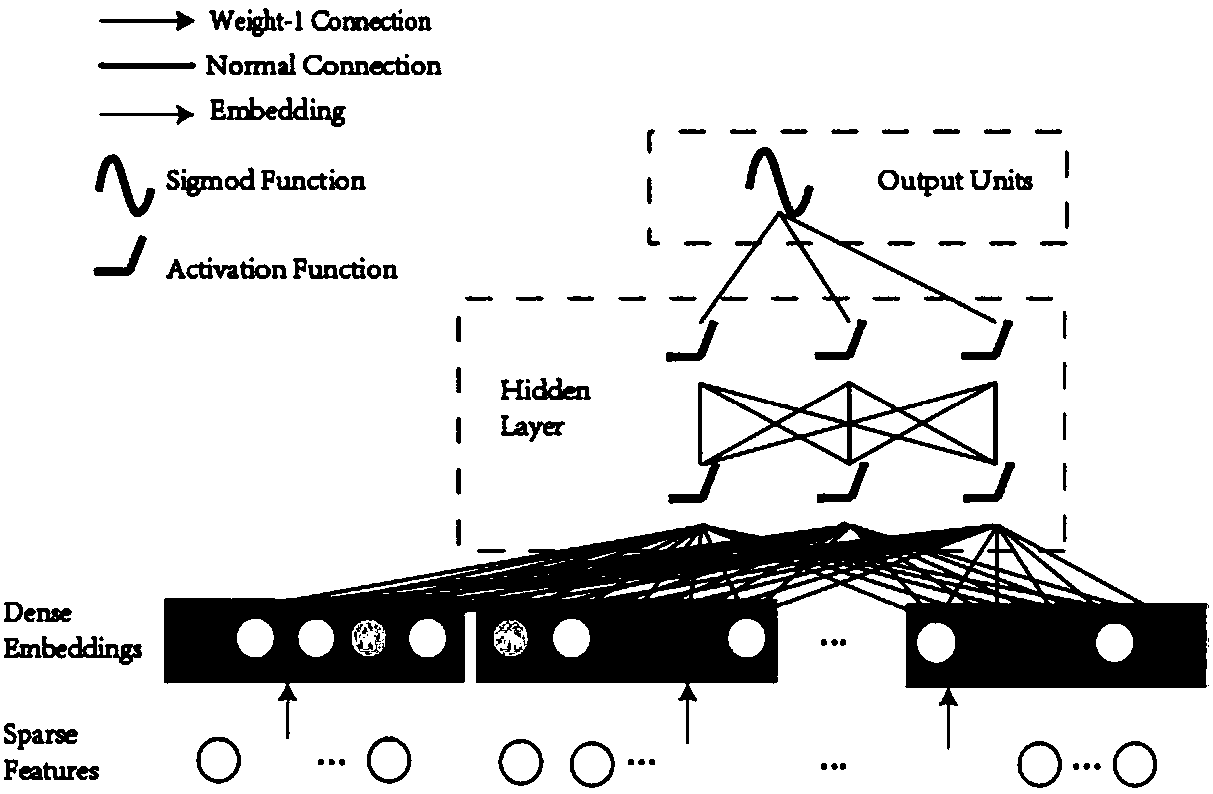
FM-N-DNN-based drug and target interaction prediction method
A prediction method and target technology, applied in the field of bioinformatics, can solve problems such as cost reduction and efficiency
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0031] The prediction method provided by the present invention includes the following steps:
[0032] 1. Data acquisition
[0033] In this example, 10731 pairs of real drug-target interaction pairs were obtained from the DrugBank database, and these were marked as positive samples. For negative samples, since the number of uncertain data is much larger than known interactions, in order to alleviate the problem of unbalanced data, random downsampling is used to generate negative samples. First, drug-target pairs are divided into different drugs and targets to form independent drug groups and target groups. The numbers of drugs and targets were 4292 and 2311, respectively. Next, drugs and targets are randomly selected to construct new drug-target pairs. Then, drug-target pairs already in the positive set are replaced with other non-redundant pairs. Finally, these drug-target pairs are labeled as negative samples. This results in a dataset of 22,719 negative samples, about t...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com