Nucleobase editors and uses thereof
A technology of amino acids and nucleotides, applied in nucleic acid vectors, genetic engineering, chemical instruments and methods, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0456] Embodiment 1: Cas9 deaminase fusion protein
[0457] A number of Cas9:deaminase fusion proteins were generated and the resulting fusions were characterized for deaminase activity. The following deaminases were tested:
[0458] Human AID (hAID):
[0459] MDSLLMNRRKFLYQFKNVRWAKGRRETYLCYVVKRRDSATSFSLDFGYLRNKNGCHVELLFLRYISDWDLDPGRCYRVTWFTSWSPCYDCARHVADFLRGNPYLSLRIFTARLYFCEDRKAEPEGLRRLHRAGVQIAIMTFKDYFYCWNTFVENHERTFKAWEGLHENSVRLSRQLRRILLPLYEVDDLRDAFRTLGLLD(SEQ ID NO:607)
[0460] Human AID-DC (hAID-DC, a truncated form of hAID with 7-fold increased activity):
[0461]MDSLLMNRRKFLYQFKNVRWAKGRRETYLCYVVKRRDSATSFSLDFGYLRNKNGCHVELLFLRYISDWDLDPGRCYRVTWFTSWSPCYDCARHVADFLRGNPNLSLRIFTARLYFCEDRKAEPEGLRRLHRAGVQIAIMTFKDYFYCWNTFVENHERTFKAWEGLHENSVRLSRQLR:(60NOGLHENSVRLSRQLR)
[0462] Rat APOBEC1 (rAPOBEC1):
[0463] MSSETGPVAVDPTLRRRIEPHEFEVFFDPRELRKETCLLYEINWGGRHSIWRHTSQNTNKHVEVNFIEKFTTERYFCPNTRCSITWFLSWSPCGECSRAITEFLSRYPHVTLFIYIARLYHHADPRNRQGLRDLISSGVTIQIMTEQESGYCWRNFVNYSPSNEAHWPRYP...
Embodiment 2
[0499] Example 2: Deamination of DNA Target Sequences
[0500] Exemplary deamination targets. The dCas9:deaminase fusion proteins described herein can be delivered to cells in vitro or ex vivo or to a subject in vivo, and can be used to achieve C when the target nucleotide is in position 3-11 with respect to the PAM. to T or G to A conversion. Exemplary deamination targets include, but are not limited to, the following: CCR5 truncation: Any codon encoding Q93, Q102, Q186, R225, W86, or Q261 of CCR5 can be deaminated to generate a stop codon, which results in CCR5 The non-functional truncation of has application in HIV treatment. APOE4 mutation: The mutant codons encoding the C11R and C57R mutant APOE4 proteins can be deaminated to restore wild-type amino acids, which has applications in the treatment of Alzheimer's disease. eGFP truncation: Any codon encoding Q158, Q184, Q185 can be deaminated to generate a stop codon, or the codon encoding M1 can be deaminated to encode I,...
Embodiment 3
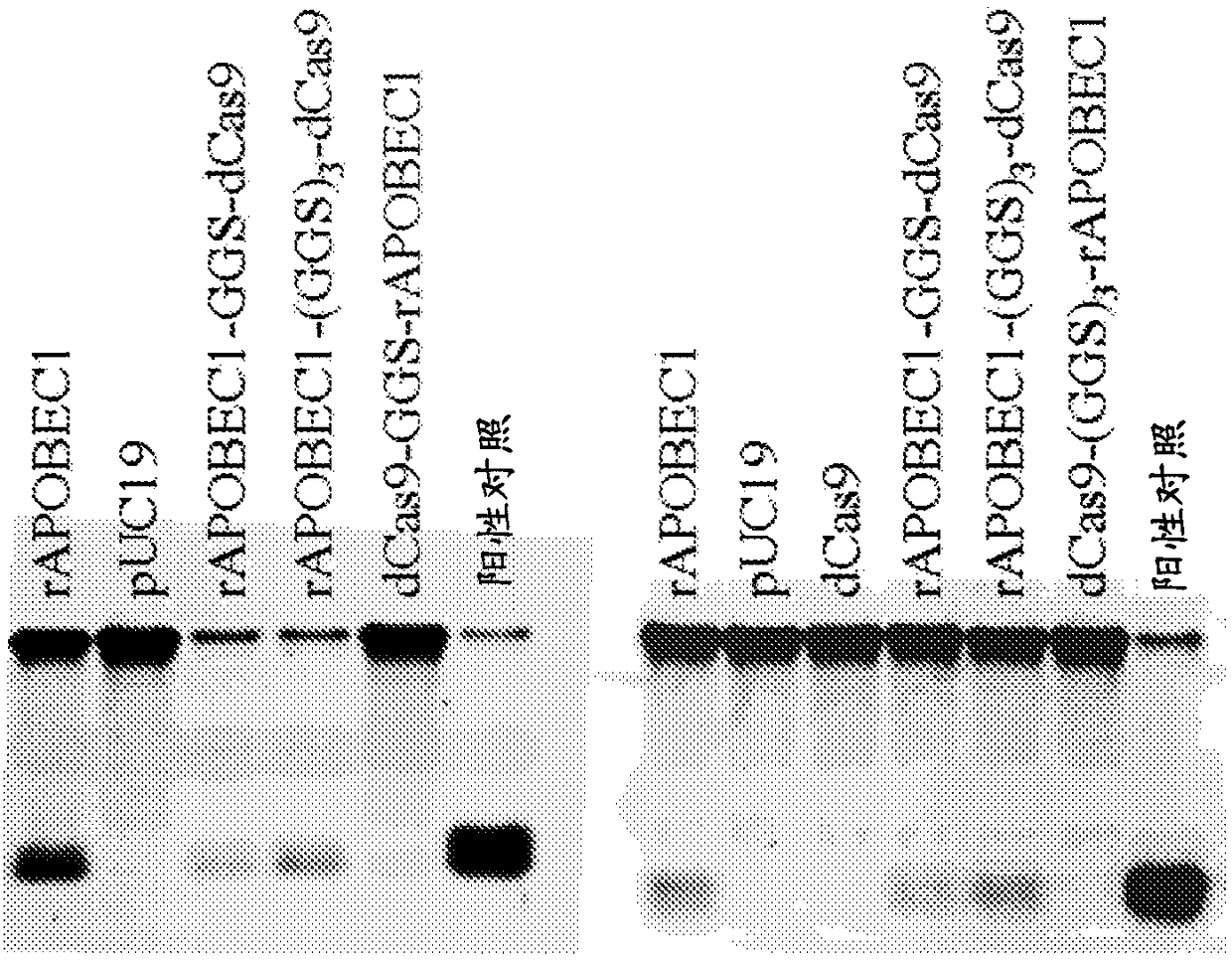
[0507] Example 3: Uracil Glycosylase Inhibitor Fusions Improve Deamination Efficiency
[0508] Directly programmable nucleobase editing efficiency in mammalian cells via dCas9:deaminase fusion proteins can be significantly improved by fusing uracil glycosylase inhibitor (UGI) to dCas9:deaminase fusion proteins.
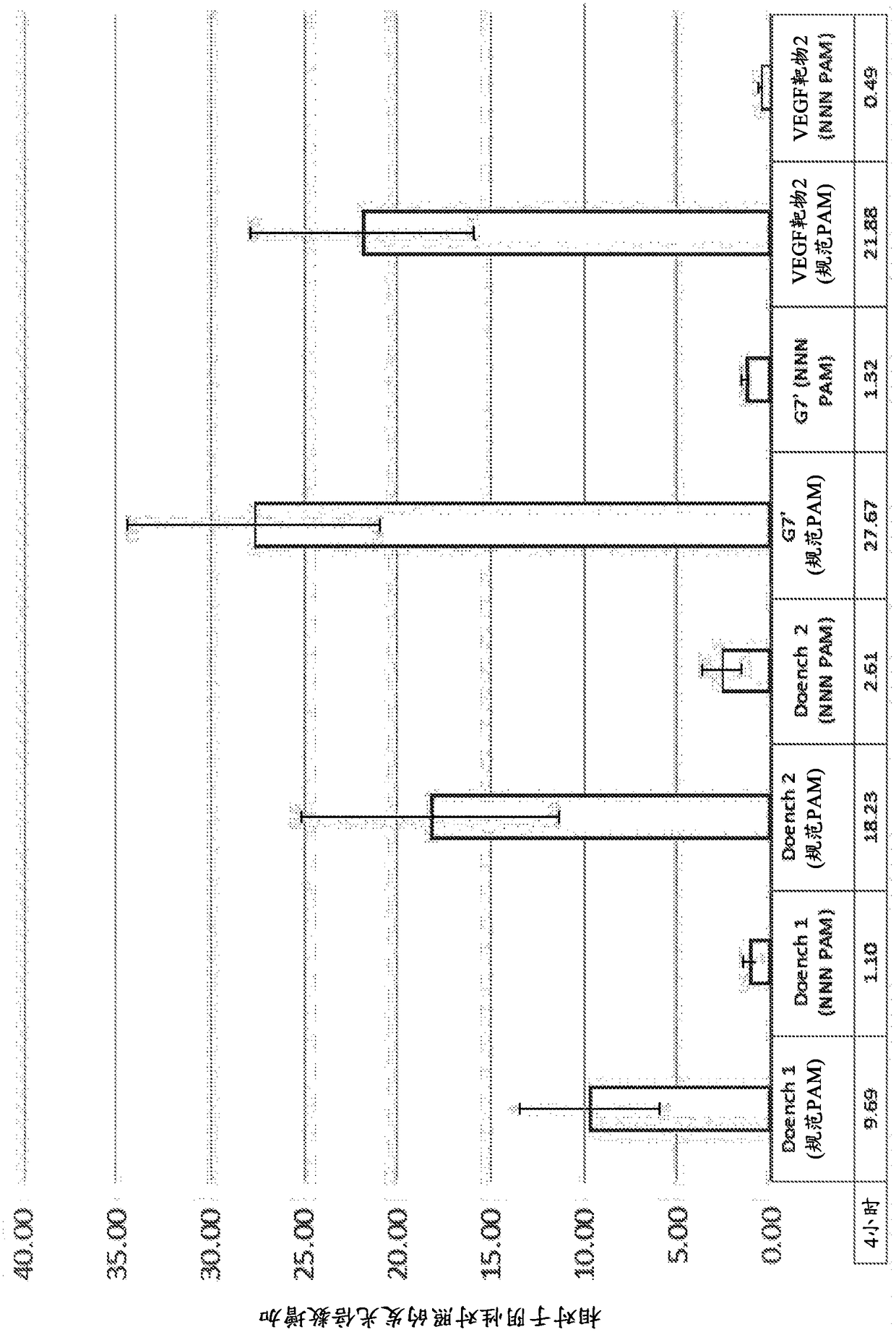
[0509] Figure 9 In vitro C→T editing efficiency in human HEK293 cells using rAPOBEC1-XTEN-dCas9 is shown:
[0510]
[0511]
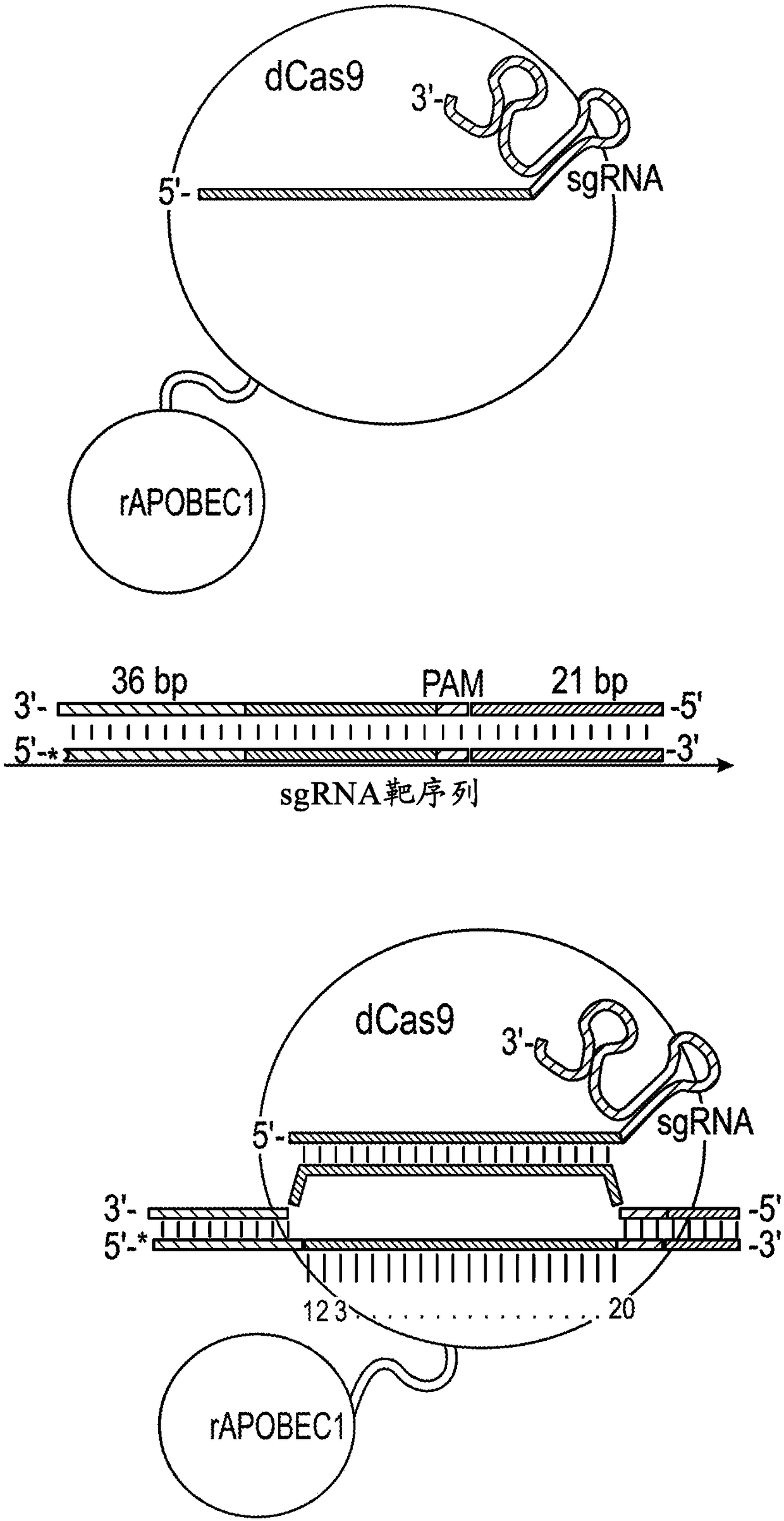
[0512] The sequence of the protospacer is as follows:
[0513]
[0514] *PAMs are boxed, C residues within the target window (positions 3-11) are numbered and bold.
[0515] Figure 10 demonstrated that the efficiency of C→T editing of the same protospacer sequence in HEK293T cells was greatly enhanced when the UGI domain was fused to the rAPOBEC1:dCas9 fusion protein.
[0516]
[0517]
[0518] From the sequencing of both strands of the target sequence it was shown that Figure 9 and Figure 10 percentage in . Since only...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Minimum inhibitory concentration | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com