Enhanced sleeping beauty transposons, kits and methods of transposition
A technology of transposon and beauty, applied in the field of nucleic acid transposition kits
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0151] result
[0152] The PAI subdomain of the SB transposase mediates primary substrate contacts
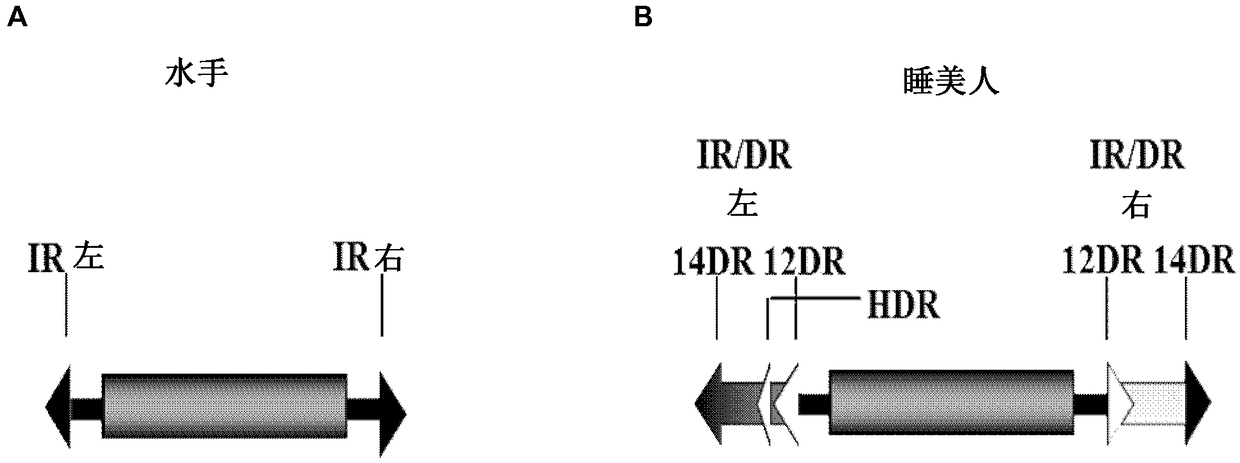
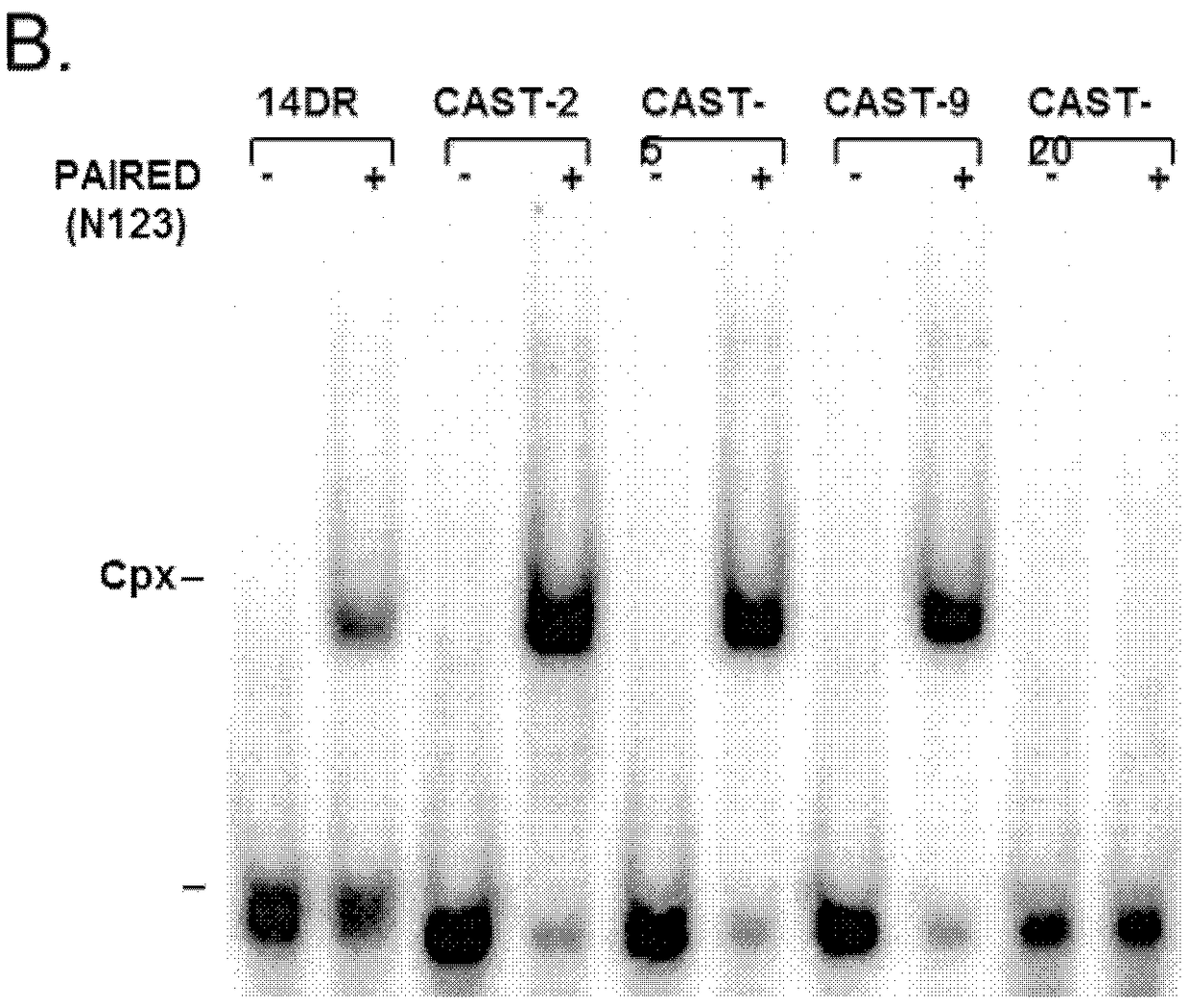
[0153]The DR of the IR / DR has a composite structure, recognized by a composite DNA-binding domain. The DNA-binding domain of the SB transposase consists of two helix-turn-helix (HTH) motifs, called PAI and RED, present in the PAX of transcription factors based on their similarity to the paired (PAIRED) domain. in the family (Izsvak Z, et al., 2002 J Biol Chem, 277:34581-8.; Czerny T, et al., 1993. Genes Dev., 7:2048-61.). Both subdomains are involved in sequence-specific DNA-binding: PAI binds to the 3'-part of the dichotomous transposase binding site represented by DR while RED interacts with its 5'-part (Izsvak Z, et al., 2002. J Biol Chem, 277:34581-8). In addition to DNA binding, PAI has previously been shown to have a protein-protein interaction interface (Izsvak Z, et al., 2002. J Biol Chem, 277:34581-8.). It is worth noting that the four DRs of SB are not homogeneous...
Embodiment 2
[0218] It has previously been demonstrated that both DNA-PKcs and ATM activity are required for efficient SB transposition (Izsvák et al., 2004, Mol Cell 13(2):279-90). Similar to DNA-PKc and ATM, ATR also belongs to the phosphatidylinositol 3-kinase-like kinase (PIKK) family and is involved in checkpoint signaling and repair. ATR is specifically activated by DNA damage during replication (Lupardus et al., 2002, Genes Dev 16(18):2327-32). Caffeine is an inhibitor of ATM, ATR and mTOR (also a member of PIKK), but not DNA-PKC (Sarkaria et al., 1999, Cancer Res. 59(17):4375-82). The inventors detected SB transposition using a standard transposition assay under caffeine treatment (4 mM).
[0219] Compared with the control, the transposition frequency was reduced by about 50% after caffeine treatment ( Figure 7A ). To elucidate whether efficient SB transposition is specifically required for ATR signaling, a stable TET-inducible cell line in which ATR function could be regulated...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com