A third-generation sequencing zooplankton genome DNA extraction method and application
A zooplankton and extraction method technology, applied in the field of genetic engineering, can solve the problems of spending huge manpower and material resources, unable to meet the requirements of building a library, etc., and achieve the effects of stable extraction product purity, simple method, and efficient removal of pigments
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0063] (1) Individual collection of Diaphanosoma dubium, the steps are as follows:
[0064] The daphnia spp. fuzzy uses fresh living body, and this species can achieve absolute dominance in summer (June to August) in some water bodies. Therefore, in this experiment, water samples of zooplankton were collected from a lake on a university campus in Guangzhou by using a 120 μm trawl net. In the laboratory, the water samples containing zooplankton passed through 600 μm and 250 μm mesh screens in turn. Immediately transfer the zooplankton above the 250μm mesh sieve to a 1L beaker filled with drinking purified water, in which 100u / L penicillin and 100μg / L streptomycin were added in advance to reduce pollution. After 5 to 6 hours, manually select the required fuzzy Daphnia spp. under incandescent light. In order to ensure the purity of Daphnia chevrosum, the selection was repeated 2 to 3 times, and the selected individuals were put into pure water containing the same antibiotic conc...
Embodiment 2
[0078] (1) Individual collection of three kinds of Cladocera, the steps are as follows:
[0079] Three species of Cladocera (Diaphanosoma dubium, D.dubium), Leptodora richard (L.richard) and Bythotrephes longimanus (Bythotrephes longimanus , B.longimanus)) conducted an experimental comparison; where:
[0080] The daphnia spp. fuzzy uses fresh living body, and this species can achieve absolute dominance in summer (June to August) in some water bodies. Therefore, in this experiment, water samples of zooplankton were collected from a lake on a university campus in Guangzhou by using a 120 μm trawl net. In the laboratory, the water samples containing zooplankton passed through 600 μm and 250 μm mesh screens in turn. Immediately transfer the zooplankton above the 250μm mesh sieve to a 1L beaker filled with drinking purified water, in which 100u / L penicillin and 100μg / L streptomycin were added in advance to reduce pollution. After 5 to 6 hours, manually select the required fuzzy D...
Embodiment 3
[0097] (1) Individual collection of zooplankton, the steps are as follows:
[0098] For the zooplankton, all the fresh live zooplankton collected in the water body at one time were used. In this experiment, zooplankton water samples were collected from a lake on a university campus in Guangzhou by using a 120 μm trawl net. In the laboratory, the water samples containing zooplankton were passed through a 600 μm mesh sieve to remove impurities. Immediately transfer the zooplankton in the water sample to a 1L beaker filled with drinking purified water, in which 100u / L penicillin and 100μg / L streptomycin were added in advance to reduce pollution. After 5-6 hours, the zooplankton was collected again with a 120 μm mesh sieve, and put into purified water containing the same antibiotic concentration to obtain fresh individual zooplankton.
[0099] (2) Zooplankton genome DNA extraction, the steps are as follows:
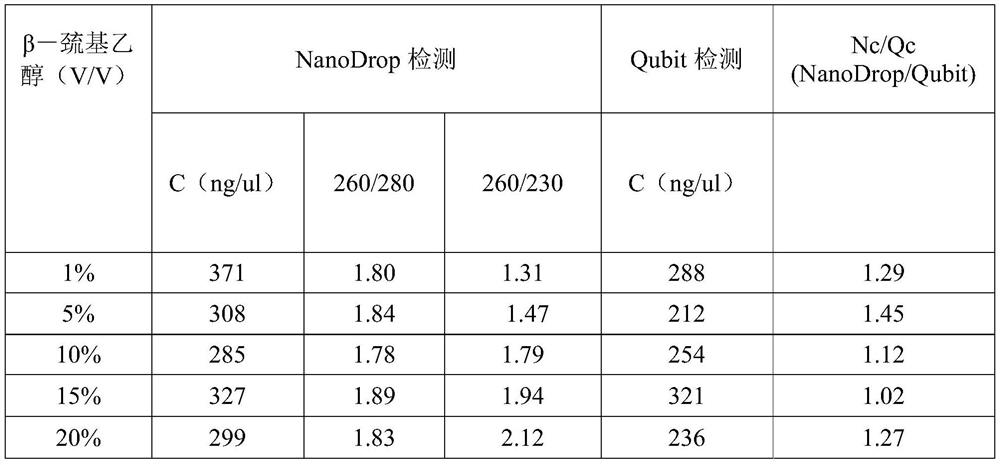
[0100] 1) Take the kit ( The buffer solution GA in the company's mar...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com