Establishing method for MSAP technical system of apocarya
A thin-shell hickory nut and a technology for establishing methods, which are applied in biochemical equipment and methods, and microbial measurement/inspection, etc., can solve problems that have not been reported in the research, and achieve the effect of low silver staining background and clear target bands
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
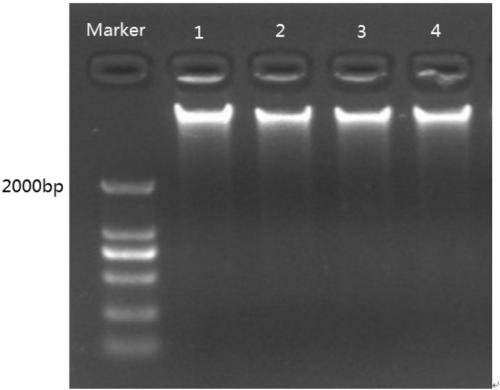
[0057] Example 1 extracts the extraction of the pecan genome DNA
[0058] Such as figure 1 As shown, samples of male flowers and leaves (the fifth pair from bottom to top) of pecan 'Pony' and 'Shaoxing' varieties were collected respectively, and genomic DNA was extracted using a kit (the extraction kit was purchased from Shanghai Shenggong Plant Genome) , wherein lanes 1, 2, 3 and 4 are the DNA samples of poni leaves, poni male flowers, Shaoxing leaves and Shaoxing male flowers, respectively. Then the quality of the extracted pecan genomic DNA was mixed, and the subsequent system optimization was carried out.
Embodiment 2
[0059] Embodiment 2 double-enzyme digestion of pecan genome DNA
[0060] Using EcoRI and MspⅠ, the double enzyme digestion reaction was carried out on the genomic DNA samples of Carya chinensis respectively. Take 6 parts of mixed DNA samples obtained in Example 1, and the enzyme digestion system is: 400ng DNA, 0.3 μL EcoR I-HF (20u / μL), 0.3 μL Msp I (20u / μL) or 0.3 μL HpaⅡ (20u / μL) , 2μL Cut smart Buffer, and make up to 20μL with sterile water.
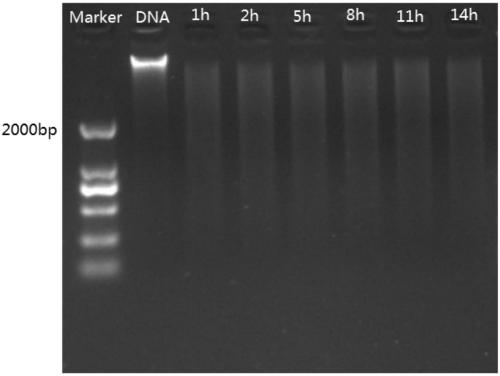
[0061] Carry out enzyme digestion in a water bath at 37°C. The enzyme digestion time is divided into 6 time gradients, namely 1h, 2h, 5h, 8h, 11h, and 14h. After each time gradient enzyme digestion is completed, put it into a PCR instrument at 65°C for 20 minutes to inactivate the enzyme. live.
[0062] figure 2 The electrophoresis of pecan genomic DNA after double digestion with Msp I. Such as figure 2 As shown, 1h, 2h, 5h, 8h, 11h and 14h are the states after digestion of the mixed DNA sample for 1h, 2h, 5h, 8h, 11h and 14h, ...
Embodiment 3
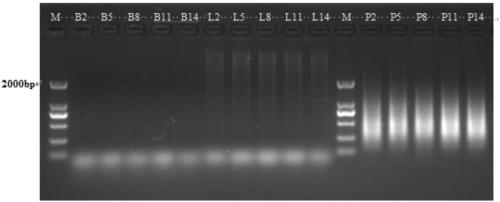
[0063] Example 3 Ligation of the double-digestion product and the linker primer
[0064] Take 5 DNA samples again from the mixed DNA sample in Example 1, apply the enzyme digestion method in Example 2 for 5 hours of enzyme digestion, and set 5 tubes of 20 μl sterile water as a blank control for 37 ° C water bath.
[0065] Ligation is performed after enzyme digestion. The system is: 20 μL of enzyme digestion product, 0.1 μl T4 (400u / μL), 2.5 μl ATP (10 mM), E adapter (mixed upstream and downstream adapters, both 50 μM) and H / M adapter (upper The downstream joints were mixed, all 50 μM) were 0.5 μL, and the sterile water was made up to 25 μL.
[0066] Add 0.5 μL E adapter and 0.5 μL H / M adapter to each tube of the control, and make up to 25 μL with sterile water. The ligation time was set to 5 gradients, respectively 2h, 5h, 8h, 11h and 14h, and the ligase was inactivated by heating at 65° C. for 10 minutes after each time gradient ligation.
[0067] Such as image 3 as shown...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com