Method for increasing compatibility of multi-PCR (Polymerase Chain Reaction) primer
A compatible and multiple technology, applied in the direction of biochemical equipment and methods, microbial measurement/inspection, etc., can solve the problems of long amplification time and difficult design of multiple PCR amplification primers, so as to reduce the amplification background and reduce the The difficulty of primer design and the effect of increasing compatibility
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0061] In this embodiment, multiplex real-time fluorescent quantitative PCR technology platform is used for detection. Considering the channel limitation of the multiplex real-time fluorescent quantitative PCR technology platform itself, all primers are used in this embodiment for multiplex amplification, and only three indicators are selected for detection during detection.
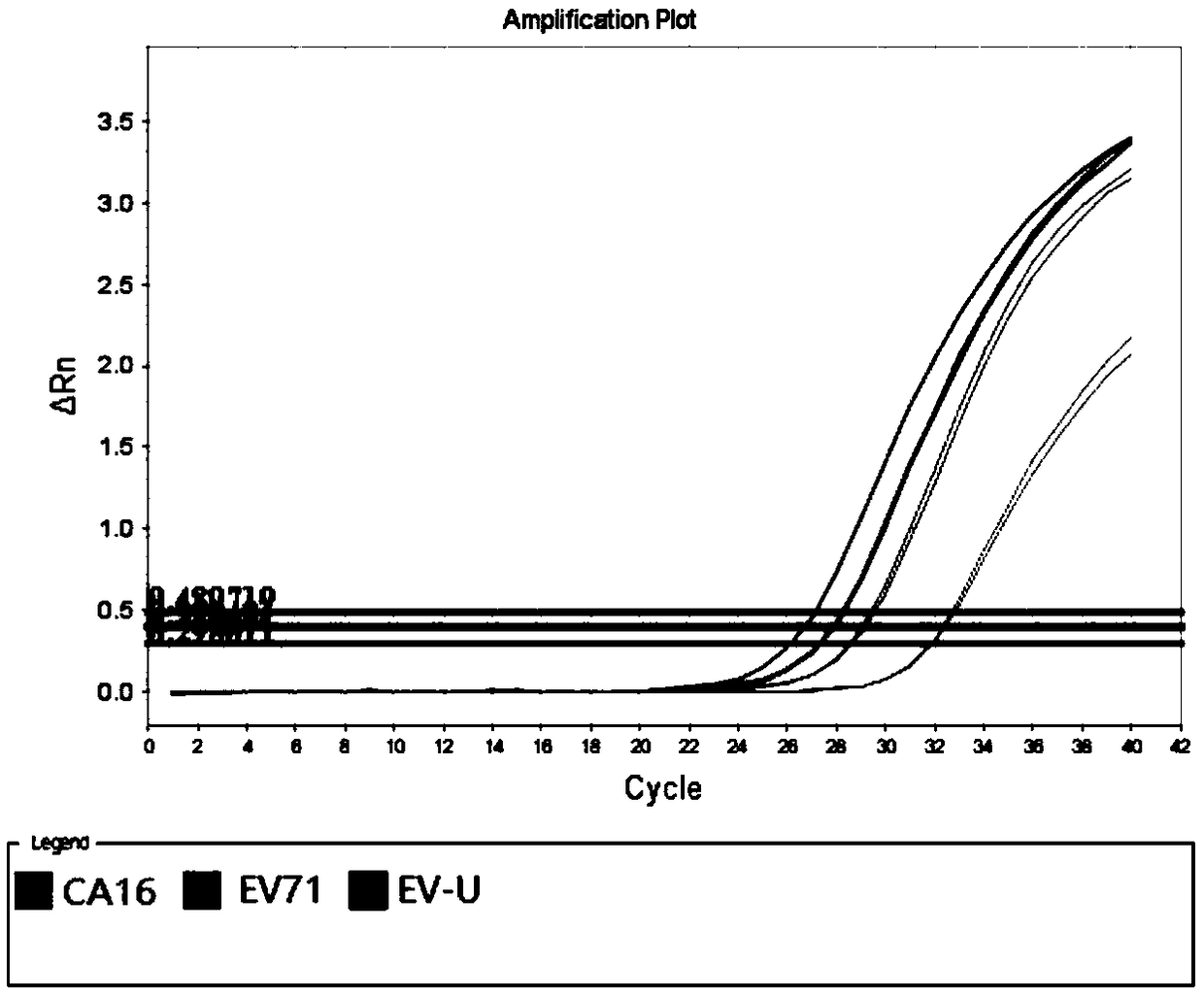
[0062] With the No. 001 / 002 / 003 sample described in Table 4 as a template, a negative control (using DEPC water as a template) was added at the same time, and the specific primers and universal primers for all pathogens described in Table 2 were used as amplification primers. The amplification method provided by Mpc-PCR was invented for multiple amplification, and the probes of EV-U, EV71, and CA16 were used as detection probes (wherein the 5' of the EV-U detection probe was labeled with FAM fluorescence, and the 3' end was subjected to BHQ -1 fluorescent labeling; EV71 detection probe 5' is HEX fluoresc...
Embodiment 2
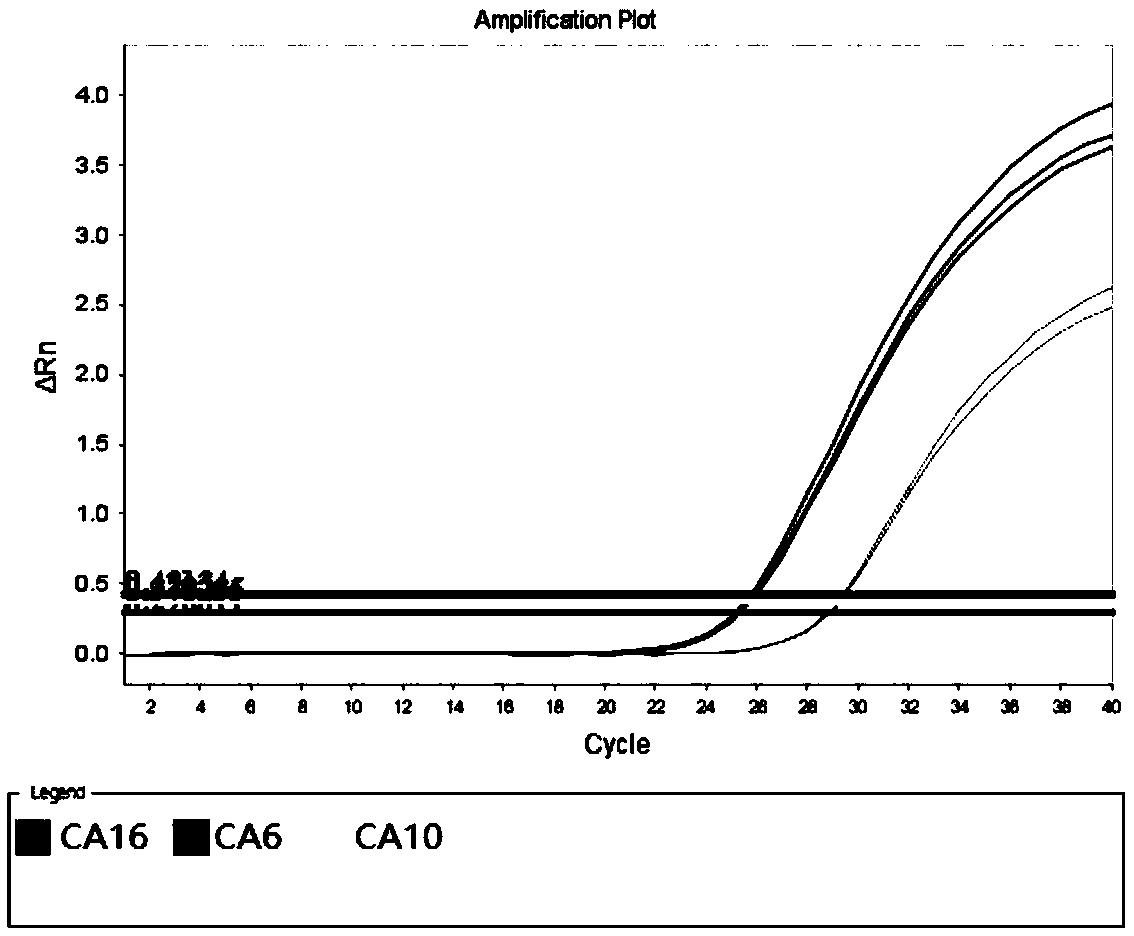
[0086] With No. 002 / 005 / 006 sample RNA described in Table 4 as a template, a negative control (using DEPC water as a template) was added simultaneously, and the specific primers and universal primers of all pathogens described in Table 2 were used as amplification primers for multiple amplification. In addition, the probes of CA6, CA10, and CA16 in Table 2 are used as detection probes (the 5' of the CA6 detection probe is fluorescently labeled with FAM, and the 3' end is fluorescently labeled with BHQ-1; the 5' of the detection probe of CA10 is fluorescently labeled with HEX labeling, BHQ-1 fluorescent labeling at the 3' end; Cy5 fluorescent labeling at the 5' end of the CA16 detection probe, and BHQ-2 fluorescent labeling at the 3' end), the multiplex fluorescent quantitative PCR technology platform was used as the detection platform for detection.
[0087] The present embodiment is carried out according to the following steps:
[0088] 1. The first step of PCR amplification ...
Embodiment 3
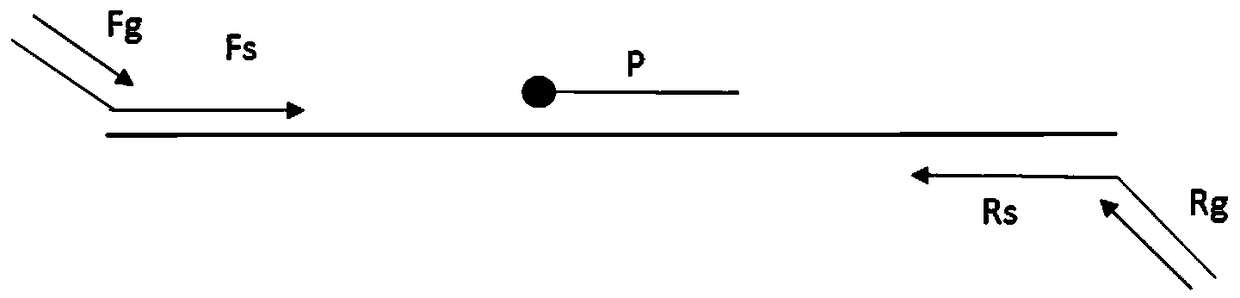
[0105] In order to test the sensitivity of this detection method, in this embodiment, the RNA of sample No. 001 / 002 / 003 / 004 / 005 / 006 described in Table 4 was used as a template, and a negative control (using DEPC water as a template) was added for detection. For each sample RNA, prepare five gradients: 100, 10 -1 、10 -2 、10 -3 、10 -4 . In another set of comparative experiments, the ratio of universal primer Rg:Fg was increased to observe the change of its detection sensitivity.
[0106] The specific primers and universal primers of all pathogens described in Table 2 were used as amplification primers for multiple amplification (wherein the reverse universal primer Rg was synthesized and the 5' end was labeled with biotin), and the specific primers of all pathogens described in Table 2 were used. The sex probe is the detection probe, and the liquid chip technology platform is used as the detection platform for detection.
[0107] The principle of liquid-phase chip technolog...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com