A Deletion Variation Identification and Clonal Counting Method for Complex Clonal Structure of Tumors
A technology of mutation identification and counting method, applied in the field of data science, to achieve the effect of reducing execution steps, improving recognition ability, and improving algorithm efficiency
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
[0058] The present invention will be described in further detail below in conjunction with the accompanying drawings, which are explanations rather than limitations of the present invention.
[0059] The present invention is a scientific and technological achievement of the following 2018 Liaoning Provincial Natural Science Foundation Project:
[0060] (1) Project number: 20180550161, project name "Tumor clone pattern detection and mutation susceptibility correlation analysis method research";
[0061] (2) Project number: 20180550855, project name "Research on the mechanism of MicroRNA-223-3 regulating Parkinson's neuroinflammation".
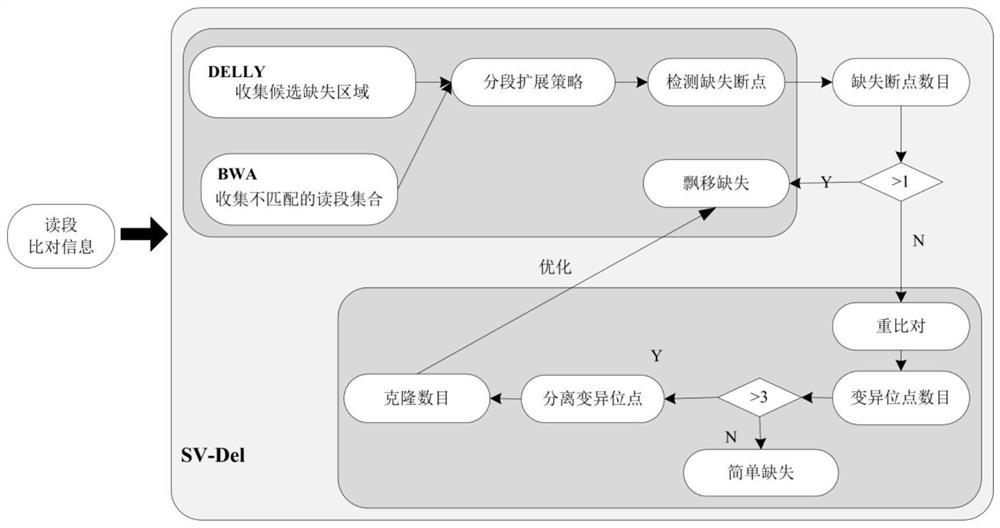
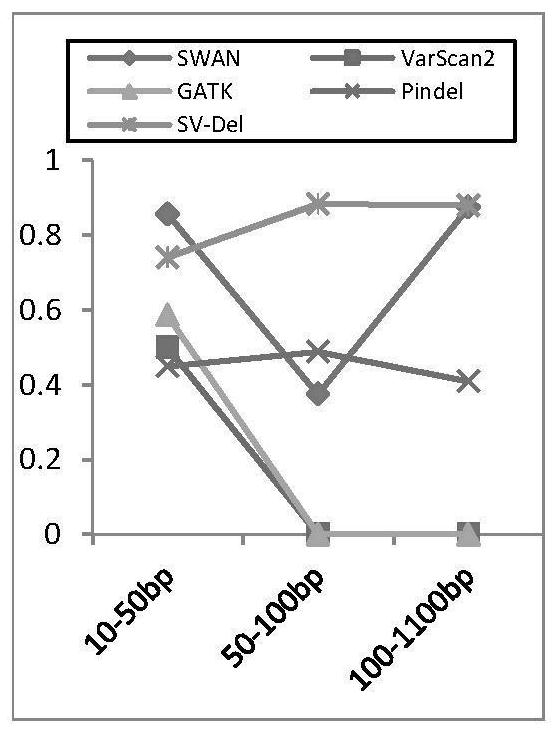
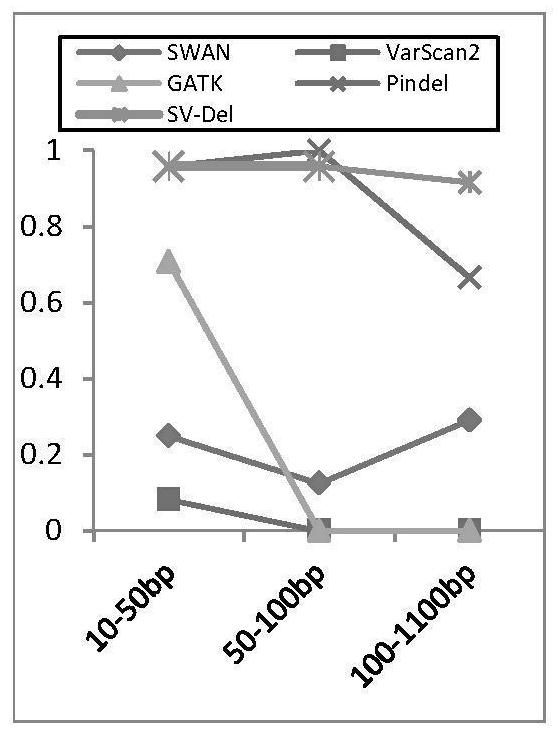
[0062] Based on high-throughput next-generation sequencing data, the present invention proposes a heuristic identification method for deletion mutations aimed at complex clonal structures in tumors. In order to improve the efficiency of the algorithm, a deletion identification method (such as the DELLY method) is first used to determine the ext...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com