A biosensor for detecting uracil-DNA glycosylase and its preparation method
A biosensor and glycosylase technology, applied in the field of biosensors, can solve the problems of difficulty in generalization, complexity, time-consuming sensitivity, etc., and achieve the effects of reducing complexity, fast reaction speed, and sensitive detection
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
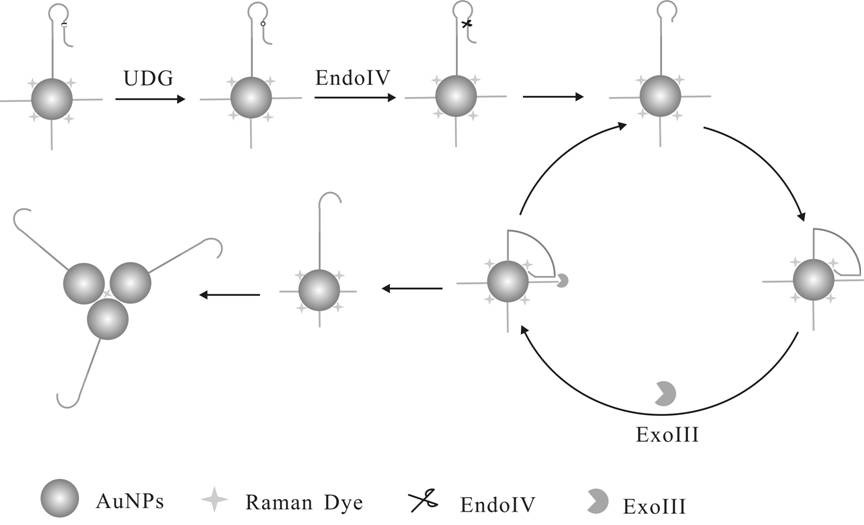
[0045] The steps for modifying Hairpin Probe and Track DNA to the surface of gold nanoparticles are as follows:
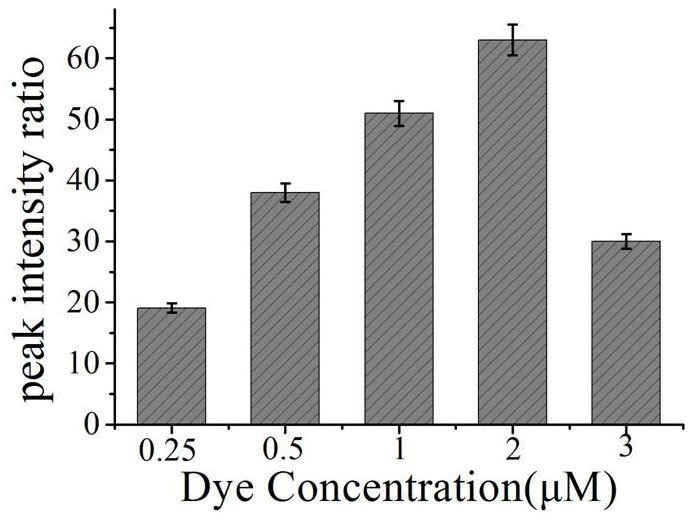
[0046] a. Take 1 mL nano-gold solution in a centrifuge tube, centrifuge for 15 min, and centrifuge the two tubes at the same time for later use. Centrifuge until the supernatant is colorless and transparent, remove the supernatant, and add 300 μL of sterile water to concentrate the nano-gold solution to 1 nM. Transfer to a 1 mL glass bottle, seal it with tin foil, and add different volumes of Raman dyes (4NTP final concentrations are 0.25uM, 0.5 uM, 1uM, 2 uM and 3 uM, respectively).
[0047] b. After standing at room temperature for 30 min, add 150 μL of -SH-modified substrate probe (Hairpin Probe and Track DNA) with a concentration of 10 μM, mix well, and place at 4 °C for 24 h.
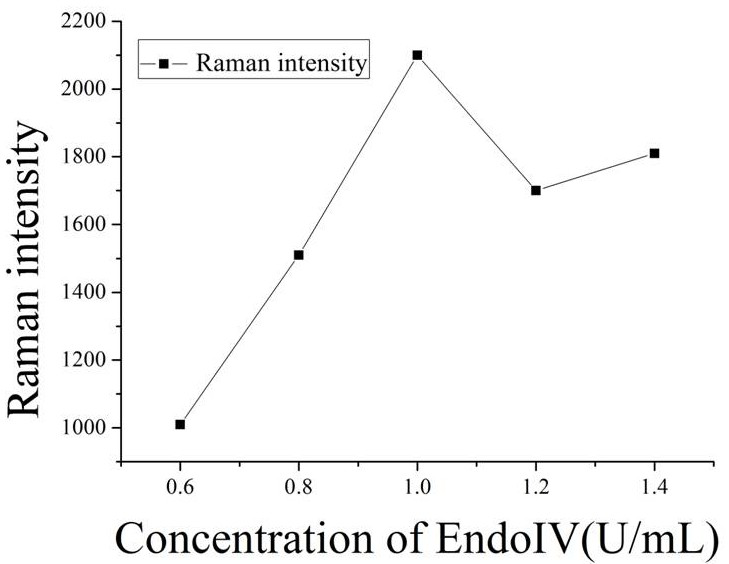
[0048] c. Slowly add 50 μL of PBS buffer several times, add magnets (soaked in aqua regia the day before) and stir for 10 min, then continue to add 27 μL of PBS buffer. Take out the m...
Embodiment 2
[0055] The steps for modifying Hairpin Probe and Track DNA to the surface of gold nanoparticles are as follows:
[0056] a. Take 1 mL nano-gold solution in a centrifuge tube, centrifuge for 15 min, and centrifuge the two tubes at the same time for later use. Centrifuge until the supernatant is colorless and transparent, remove the supernatant, and add 300 μL of sterile water to concentrate the nano-gold solution to 1 nM. Transfer to a 1 mL glass bottle, seal it with tin foil, and add Raman dye (4NTP final concentration is 2 μM).
[0057] b. After standing at room temperature for 30 min, add 150 μL of -SH-modified substrate probe (Hairpin Probe and Track DNA) with a concentration of 10 μM, mix well, and place at 4 °C for 24 h.
[0058] c. Slowly add 50 μL of PBS buffer several times, add magnets (soaked in aqua regia the day before) and stir for 10 min, then continue to add 27 μL of PBS buffer. Take out the magnet, and place it at 4°C for 48 h.
[0059] d. After 48 hours, sl...
Embodiment 3
[0066] The steps for modifying Hairpin Probe and Track DNA to the surface of gold nanoparticles are as follows:
[0067] a. Take 1 mL nano-gold solution in a centrifuge tube, centrifuge for 15 min, and centrifuge the two tubes at the same time for later use. Centrifuge until the supernatant is colorless and transparent, remove the supernatant, and add 300 μL of sterile water to concentrate the nano-gold solution to 1 nM. Transfer to a 1 mL glass bottle, seal it with tin foil, and add Raman dye (4NTP final concentration is 2 μM).
[0068] b. After standing at room temperature for 30 min, add 150 μL of -SH-modified substrate probe (Hairpin Probe and Track DNA) with a concentration of 10 μM, mix well, and place at 4 °C for 24 h.
[0069] c. Slowly add 50 μL of PBS buffer several times, add magnets (soaked in aqua regia the day before) and stir for 10 min, then continue to add 27 μL of PBS buffer. Take out the magnet, and place it at 4°C for 48 h.
[0070] d. After 48 hours, sl...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com