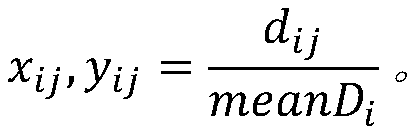Single-gene or polygene copy number detection system and method based on next generation sequencing technology
A second-generation sequencing technology and copy number technology, applied in sequence analysis, biostatistics, instruments, etc., can solve the problems of strong batch effect and low detection accuracy, and achieve the effect of less computing resources and time consumption
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0075] Example 1 Using the method of the present invention to detect single gene copy number variation
[0076] The LASSO linear regression model was trained using the data action training sets of 500 normal males and 500 normal females respectively. The positive samples to be tested include 6 samples with partial deletion or increase of DMD gene, 2 samples with partial deletion of SLC26A4 gene, and 1 sample with partial deletion of PTRRA gene. The detailed information of the sample copy number variation status is shown in Table 1, and the consistency of the 9 test samples reached 100%.
[0077] Table 1
[0078]
[0079]
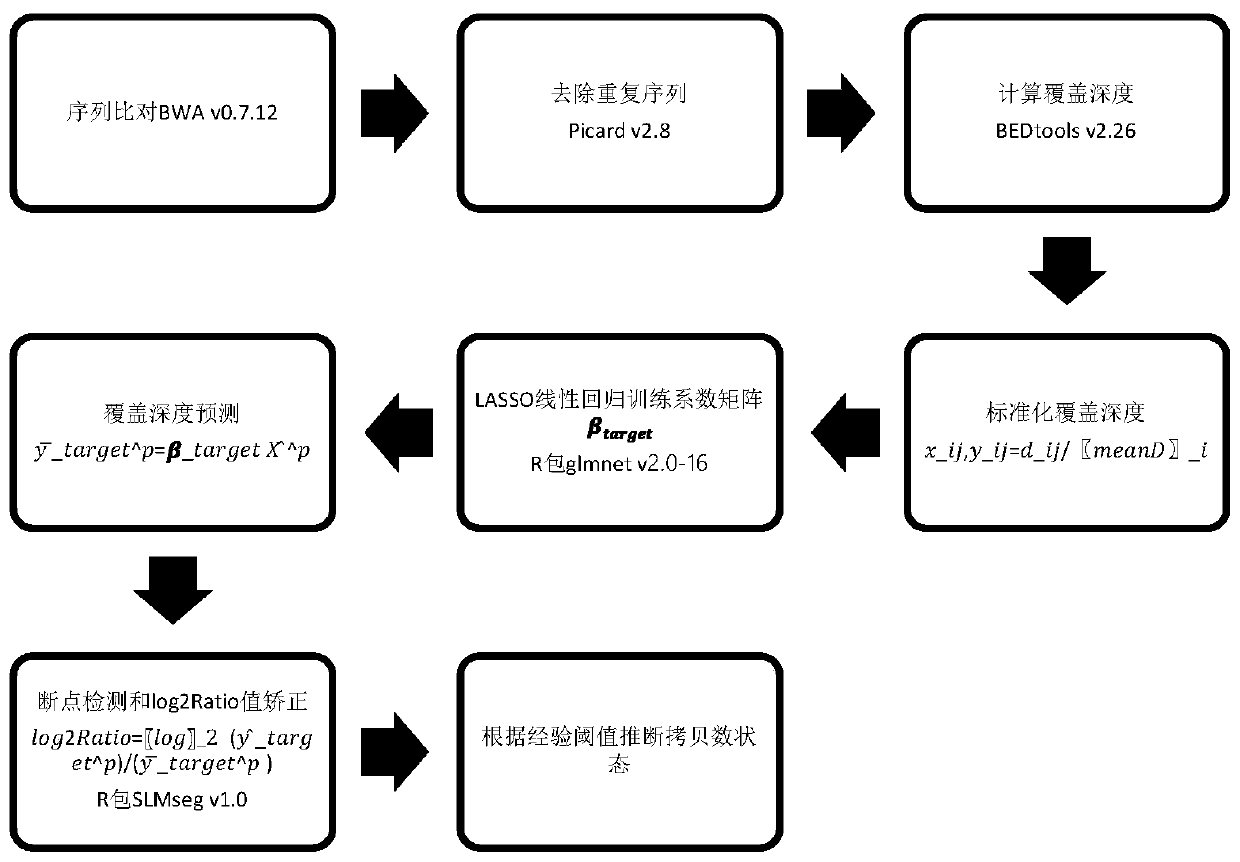
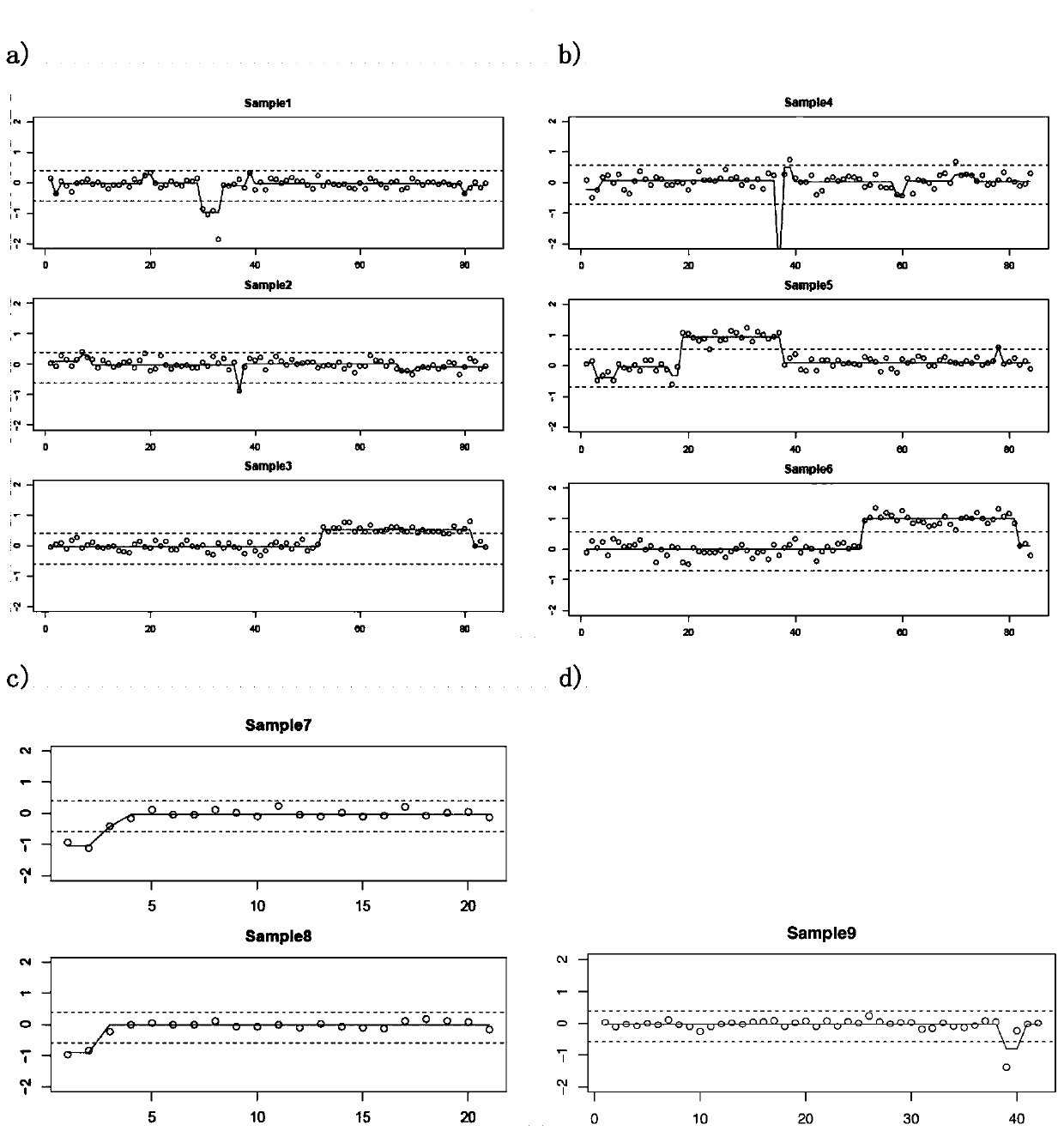
[0080] Test results such as figure 2 as shown, figure 2 The abscissa in the middle indicates the division of the divided capture interval, the ordinate indicates log2Ratio, each dot indicates the log2Ratio value of an interval, the broken line on the dot indicates the log2Ratio value after correction, and the upper and lower dotted lines indicate ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com