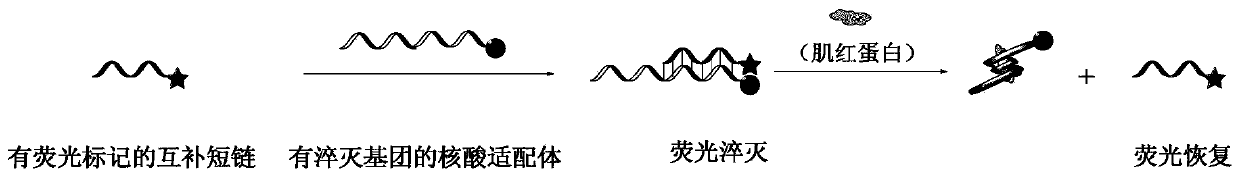
Nucleic acid aptamer fluorescence sensor for detecting myoglobin and construction method of nucleic acid aptamer fluorescence sensor
A nucleic acid aptamer and fluorescent sensor technology, which is applied in the field of analysis and detection, can solve the problems of cumbersome time-consuming, cumbersome separation and washing steps, and high cost, and achieve the effects of improving accuracy and sensitivity, shortening fluorescence recovery time, and fewer detection steps
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0042] 1. The base series structure of the nucleic acid aptamer with a quenching group is:
[0043] 5'-CCCTCCTTTCCTTCGACGTAGATCTGCTGCGTTGTTCCGA-
[0044] DABCYL-3', wherein DABCYL is a quenching group, and its molecular structure is: 4-[4-(dimethylamino)phenylazo]benzoic acid.
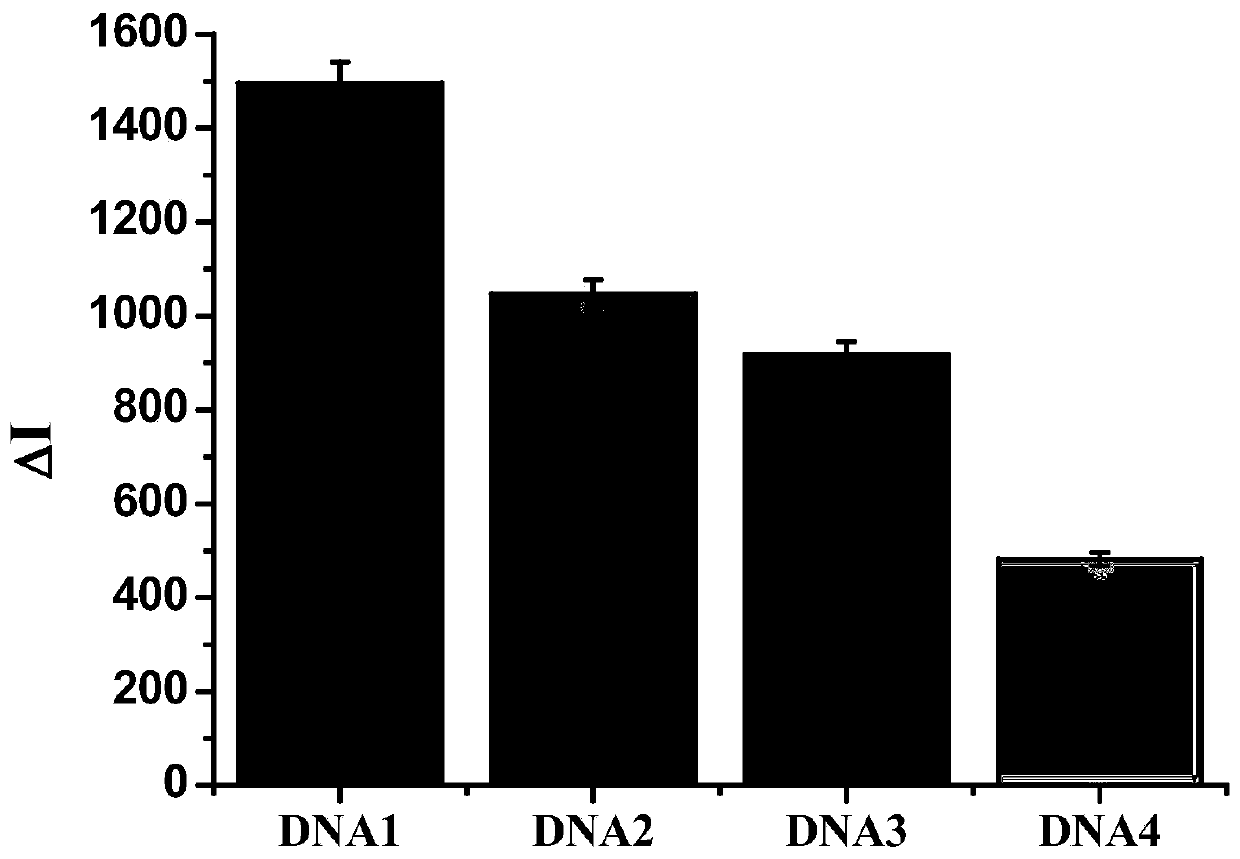
[0045] The base series structure of the complementary short chain with a fluorescent group is: DNA1: 5'-FAM-GGAACAACGC-3'; DNA2: 5'-FAM-TCGGAACAAC-3'; DNA3: 5'-FAM-CGGAACAACG-3 '; DNA4: 5'-FAM-GAACAACGCA-3', wherein FAM is a fluorescent group, and its molecular structure is: 6-carboxyfluorescein.
[0046] 2. Add 1760 μL of 0.01M PBS buffer solution (pH=7.4), 100 μL of 500 nM complementary short chain with fluorescent group and 100 μL of 500 nM nucleic acid aptamer with quencher group into a 5 mL plastic tube in sequence. The final concentrations of the complementary short chains linked with fluorescent groups and the nucleic acid aptamers linked with quenching groups were both 25 nM. The mixed solut...
Embodiment 2
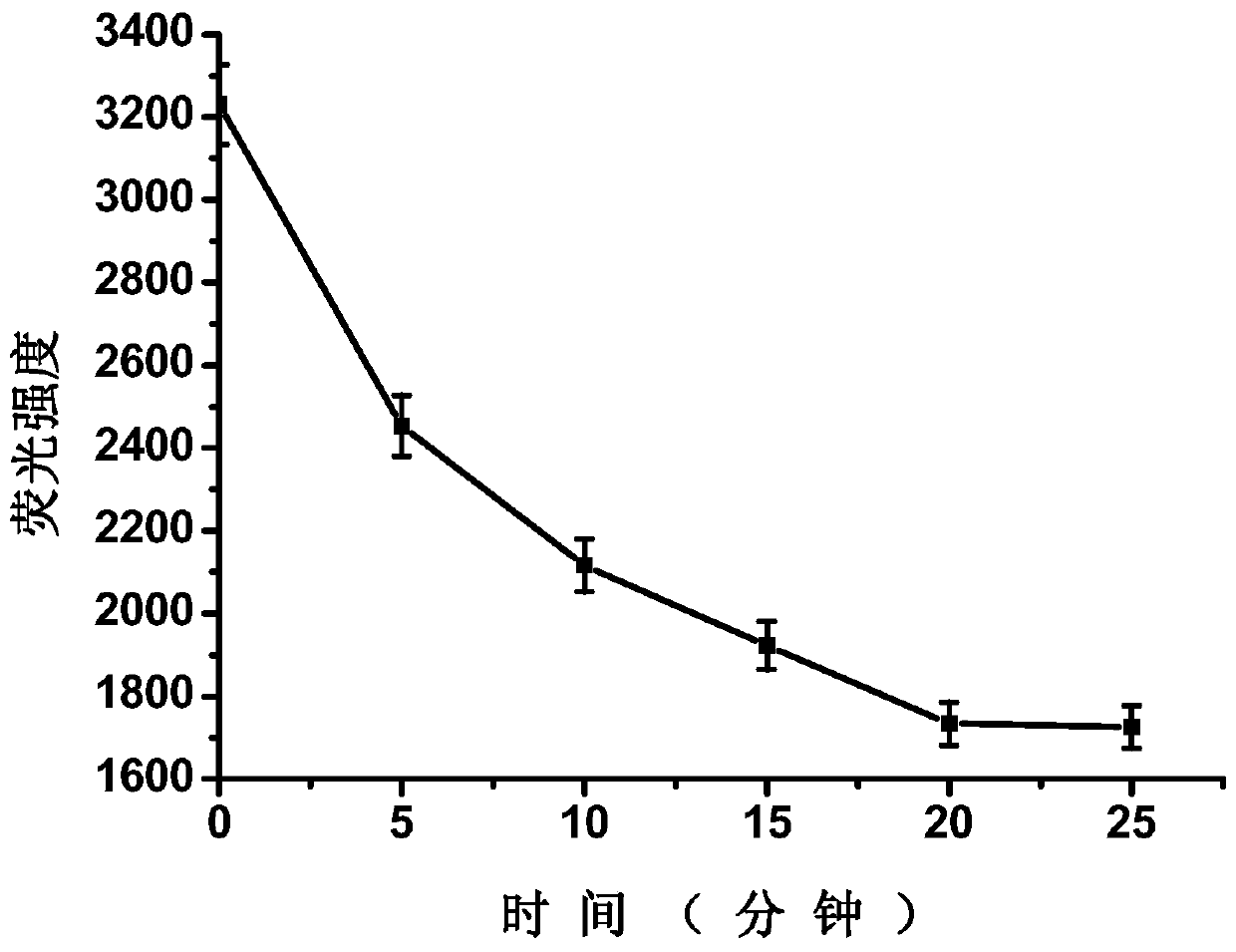
[0060] Example 2: Detection of human serum myoglobin samples and spiked samples
[0061] 1. First, the human serum sample (33.4ng / mL, the test result of the hospital) was diluted 6.68 times with 0.01M PBS buffer solution (pH=7.4), and then the macromolecules in the serum were filtered out with an ultrafiltration centrifuge tube (30kD) Protein, human serum myoglobin samples can be prepared.
[0062] Next, add 1760 μL of 0.01M PBS buffer solution (pH=7.4), 100 μL of 500 nM complementary short chains with fluorescent groups and 100 μL of 500 nM nucleic acid aptamers with quenching groups into a 5 mL plastic tube in sequence. The final concentrations of the complementary short chains linked with fluorescent groups and the nucleic acid aptamers linked with quenching groups were both 25 nM. The mixed solution was incubated at 25°C for 20 minutes.
[0063] Then, 1960 μL of the mixed solution was divided into two equal parts, each solution was 980 μL, and 20 μL of 0.01M PBS buffer s...
PUM
| Property | Measurement | Unit |
|---|---|---|
| wavelength | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to view more
Login to view more - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap