Nucleic acid reagents, kits, systems and methods for detecting Escherichia coli and Klebsiella pneumoniae and their toxicity and drug resistance
A technology for Klebsiella pneumoniae and Escherichia coli, applied in biochemical equipment and methods, microbial determination/inspection, DNA/RNA fragments, etc., can solve the problem of poor detection accuracy, time-consuming and laborious, Klebsiella pneumoniae Fewer virulence factor detection methods, etc., to achieve the effect of saving time, high conservation and specificity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
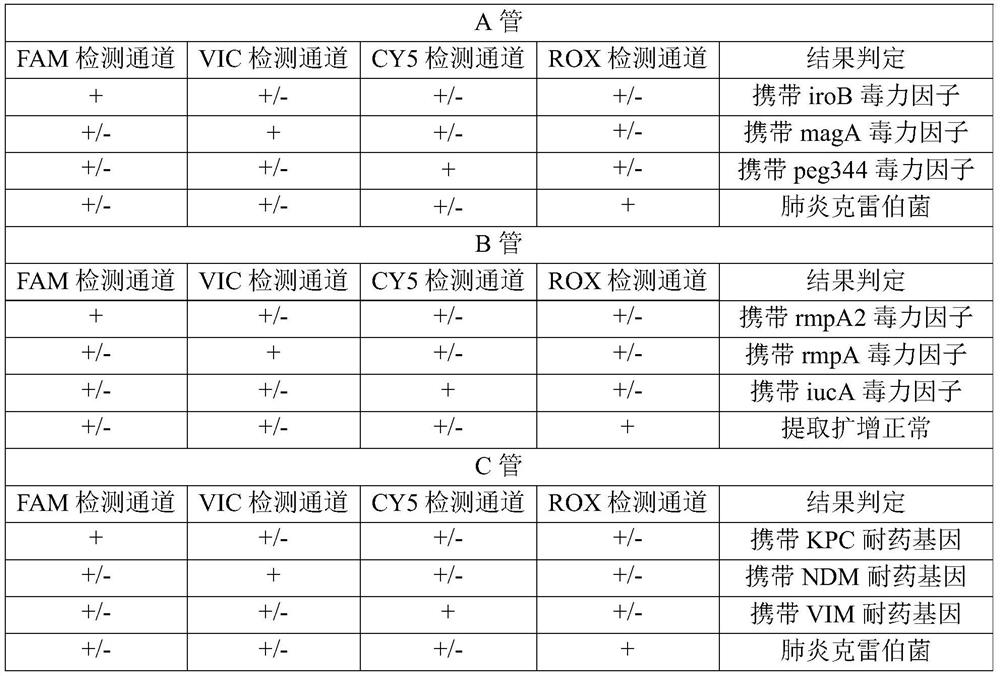
[0075] Embodiment 1 detection method and detection result judgment
[0076] 1. Primer and probe synthesis
[0077] Sequence synthesis was performed according to the primer sequences shown in Table 1 and the probe sequences shown in Table 2. In the probe, FAM is 6-carboxyfluorescein, HEX is hexachloro-6-methylfluorescein, CY5 is 5H-indocyanine, ROX is 6-carboxy-X-rhodamine, and VIC is a dye purchased from ABI , BHQ1, BHQ2 and BHQ3 are quenching groups.
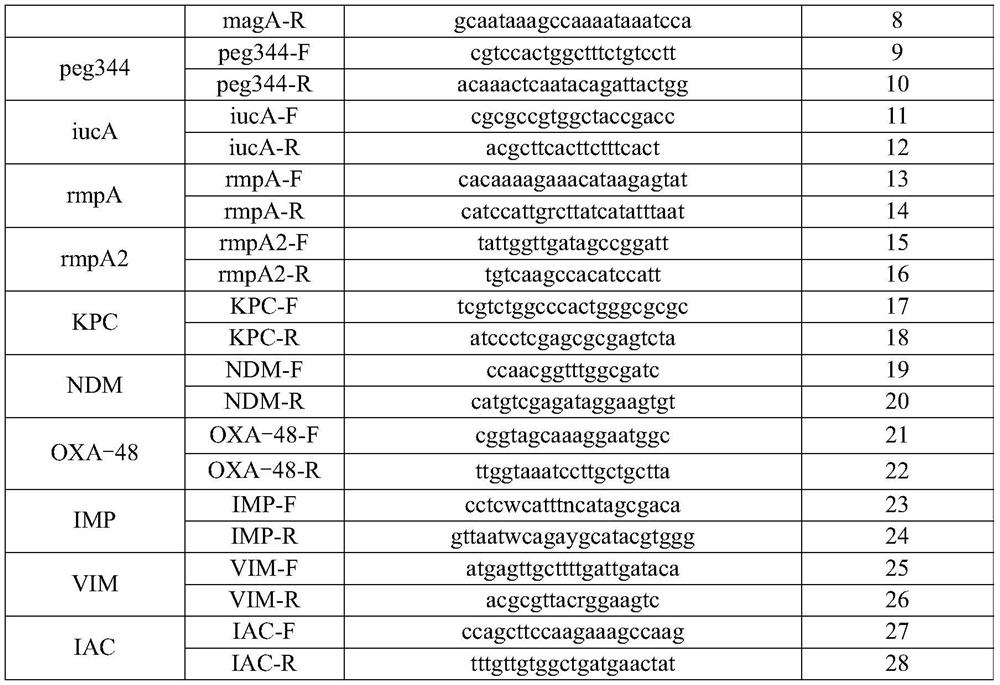
[0078] Table 1
[0079]
[0080]
[0081] Table 2
[0082] detection target probe sequence probe sequence SEQ ID NO Klebsiella pneumoniae KP-P ROX-cctacaccagctccgaccgtaccaa-BHQ2 29 Escherichia coli uidA-P CY5-tcggcatccggtcagtggcagt-BHQ3 30 iROB iroB-P FAM-agtctcggcaccgtcaaacc-BHQ1 31 magA magA-P VIC-tcgccgcaaatacgagaagtgtagt-BHQ1 32 peg344 peg344-P CY5-cctccgtgatgaggatgaacgaaagtgaag-BHQ3 33 iucA iucA-P CY5-ctatactgcgccaccagccgcgacatgat-BHQ3 34 ...
Embodiment 2
[0104] Embodiment 2 minimum detection limit verification
[0105] Escherichia coli gene, Klebsiella pneumoniae gene, iroB virulence factor, magA virulence factor, peg344 virulence factor, iucA virulence factor, rmpA virulence factor, rmpA2 virulence factor, KPC The gene fragments of the drug resistance gene, NDM drug resistance gene, IMP drug resistance gene, VIM drug resistance gene and OXA-48 drug resistance gene were connected to the PUC57 vector, and sequenced and checked to obtain the recombinant plasmids. The construction and sequencing proofreading of the above recombinant plasmids were completed by Shanghai Sangon Biotechnology Company.
[0106] The concentrations of the recombinant plasmids containing the above target gene fragments were quantified as 10 10 copies / μL, and were serially diluted to obtain a concentration of 10 5 copies / μL, 10 4 copies / μ, 10 3 copies / μL, 10 2 copies / μL, 10 1 copies / μL, 10 0 copies / μL of test sample.
[0107] According to the dete...
Embodiment 3
[0109] Example 3 specificity verification
[0110] Enterobacter cloacae (purchased from China Medical Culture Collection Center, No. 45301), Serratia marcescens (purchased from China Medical Culture Collection Center, No. 41002), Salmonella paratyphi A (purchased from China Medical Culture Collection Center, No. Culture Collection Center, No. 50001), Salmonella paratyphi B (purchased from China Medical Culture Collection Center, No. 50004), Bacillus mycoides (purchased from China Medical Culture Collection Center, No. 32012), Wei Bacillus subtilis (purchased from China Medical Culture Collection Center, No. 32011), Bacillus subtilis (purchased from China Medical Culture Collection Center, No. 32041), Staphylococcus aureus (purchased from China Medical Culture Collection Center, No. 26003), Vibrio parahaemolyticus (purchased from China Medical Culture Collection Center, No. 21617), Listeria monocytogenes (purchased from China Medical Culture Collection Center, No. 54002), chole...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com