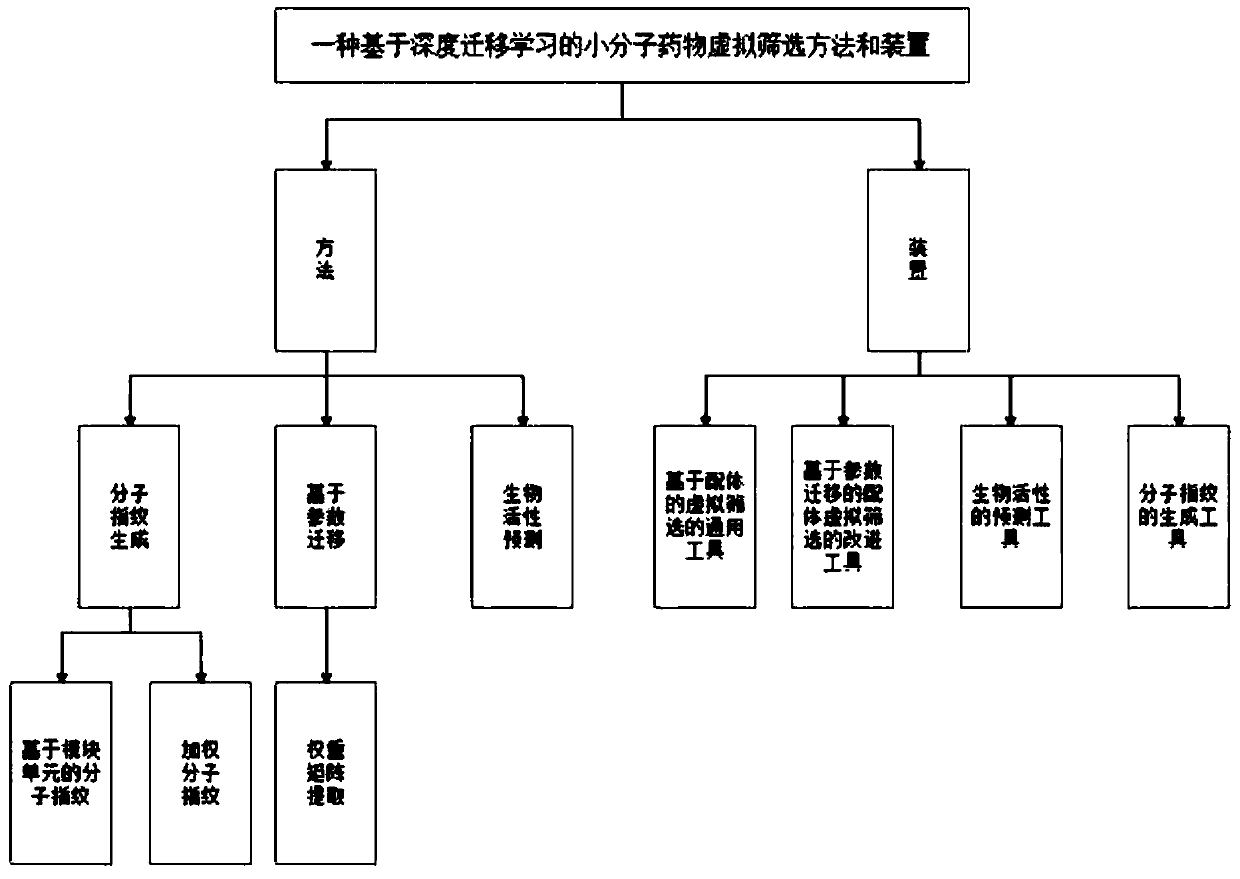
Small molecule drug virtual screening method based on deep migration learning and application thereof
A technology of virtual screening and transfer learning, which is applied in the field of virtual screening of small molecule drugs based on deep transfer learning, which can solve problems such as difficulty in obtaining useful ligand samples and insufficient information on active ligand samples.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
specific Embodiment approach
[0050] Table 1
[0051]
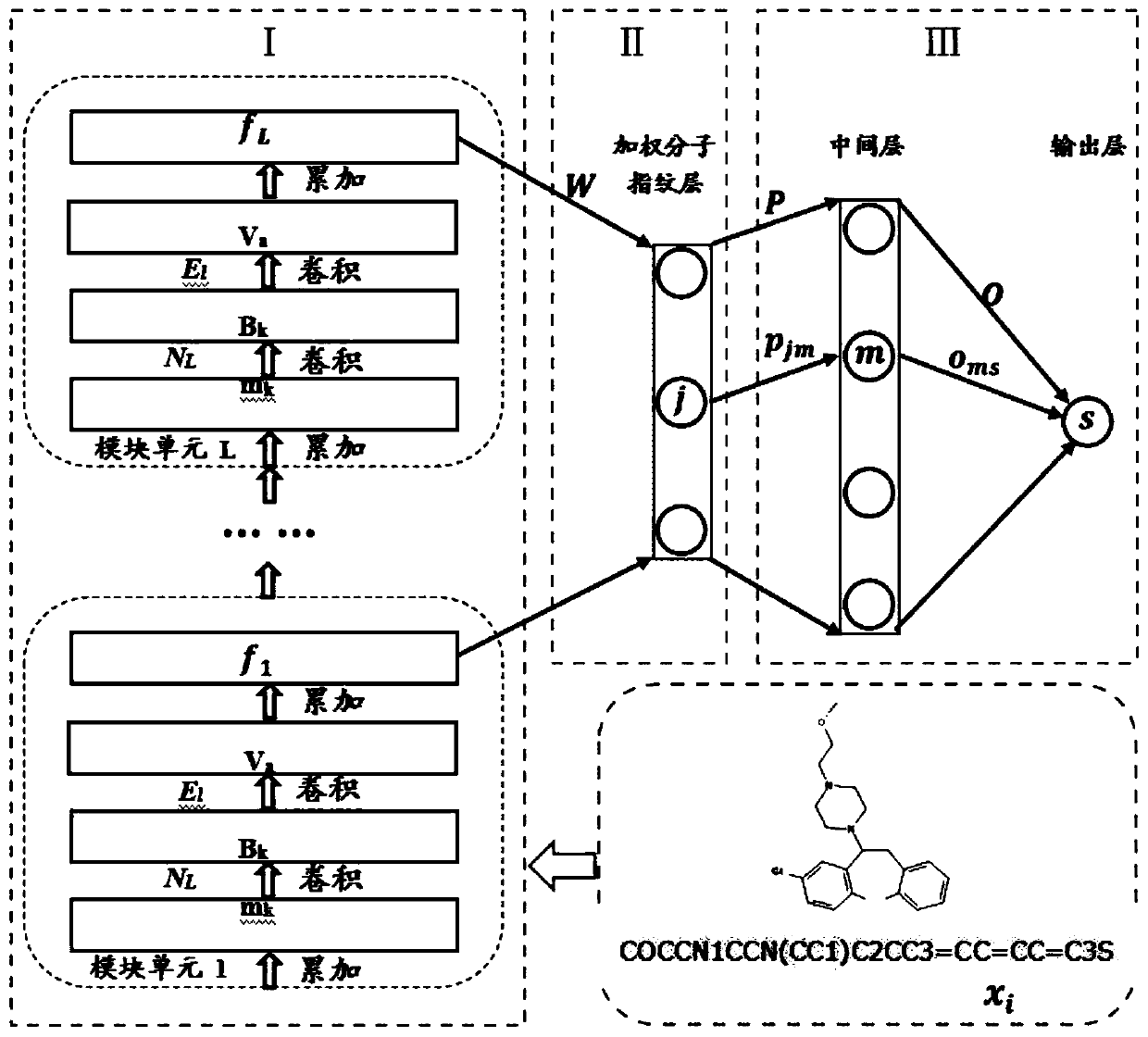
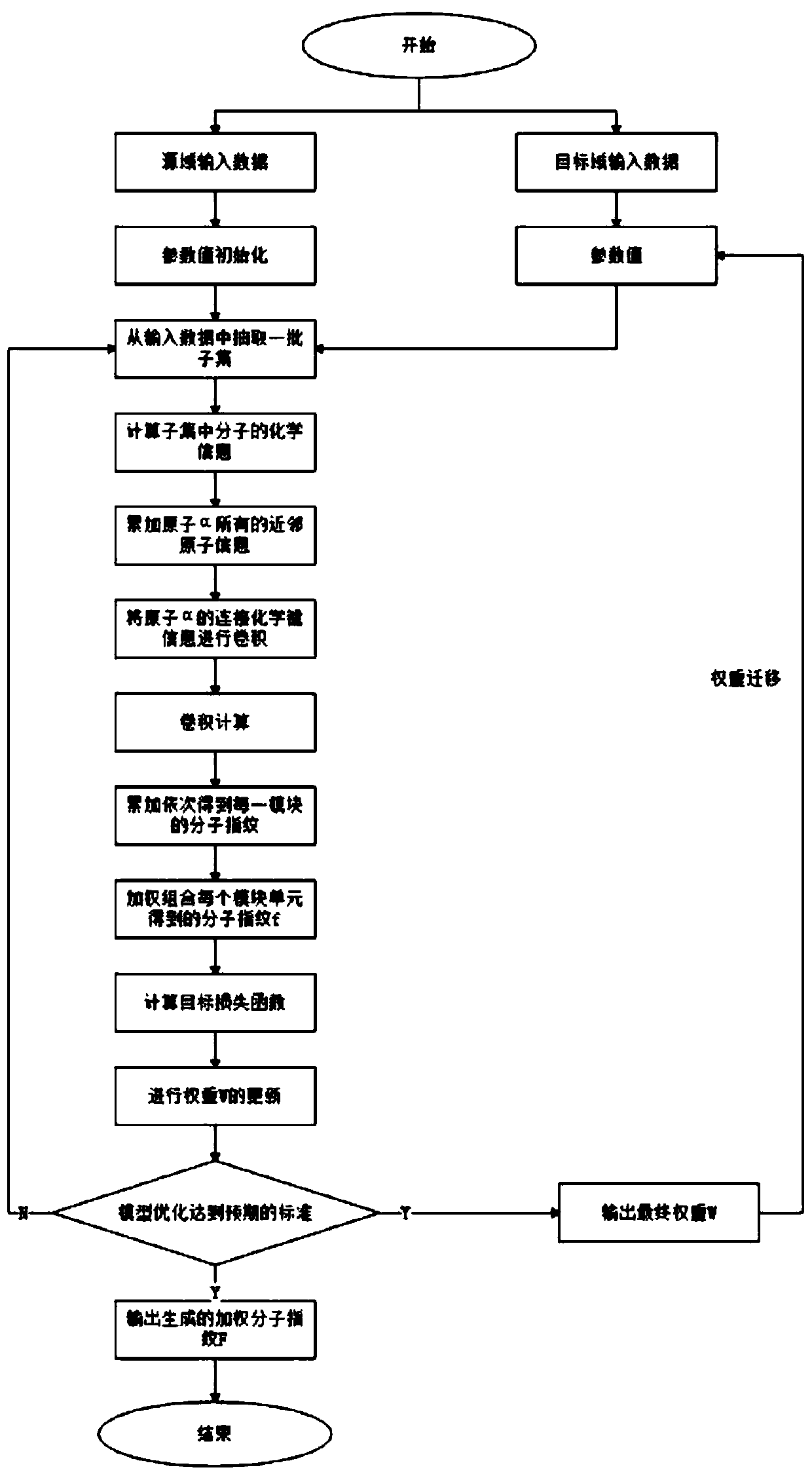
[0052] As shown in Table 1 above, this is the data set we obtained after biotechnology screening. They are grouped into one group because they all belong to homologous proteins and have a common family. Here we call it Group A, where A1-A6 It is our target domain, that is, the small sample data set we are targeting. The number of them ranges from more than one hundred to more than one thousand, which is very unfavorable for us to do deep learning, so we found our source domain again, namely AS1 , AS2, they have sample sizes in the thousands. What we have to do is to use the source domain to improve the training effect of the target domain. The specific implementation steps are as follows:
[0053] 1. Take the source domain as input and input it into our general tool demo_new1 for ligand-based virtual screening for training:
[0054] (1) Initialize network parameters, including weight matrix W, molecular fingerprint f 0 ;
[0055] (2) Randomly s...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com