Nucleic acid construct for gene editing
A nucleic acid construct and gene editing technology, applied in the biological field, can solve the problems of low single-base editing efficiency and the limitation of the editable range of plant genomes
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
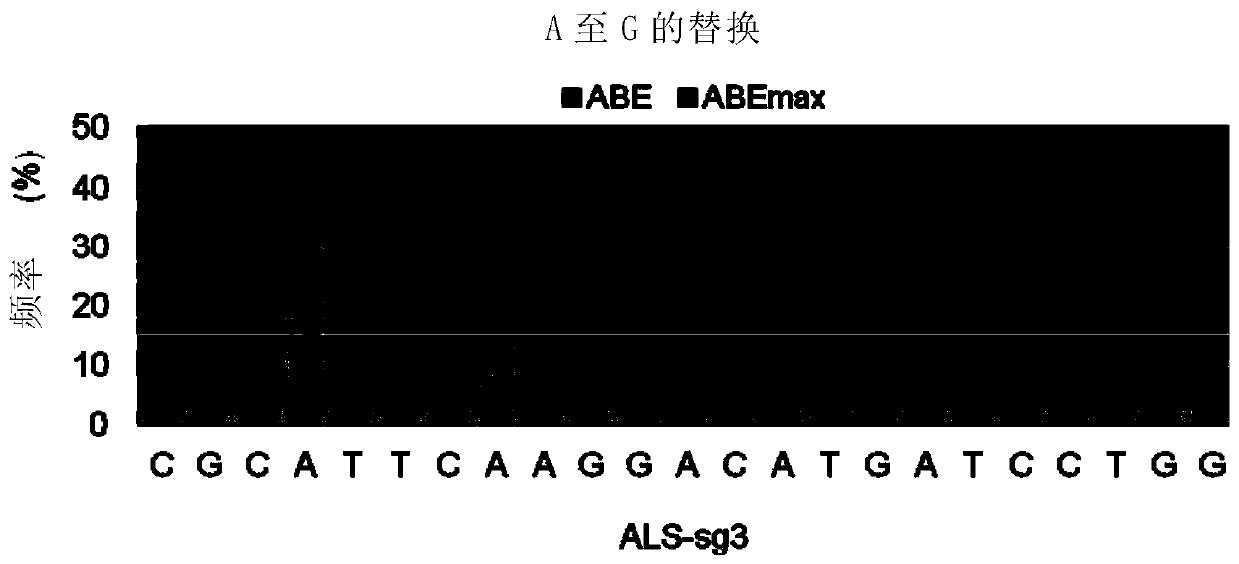
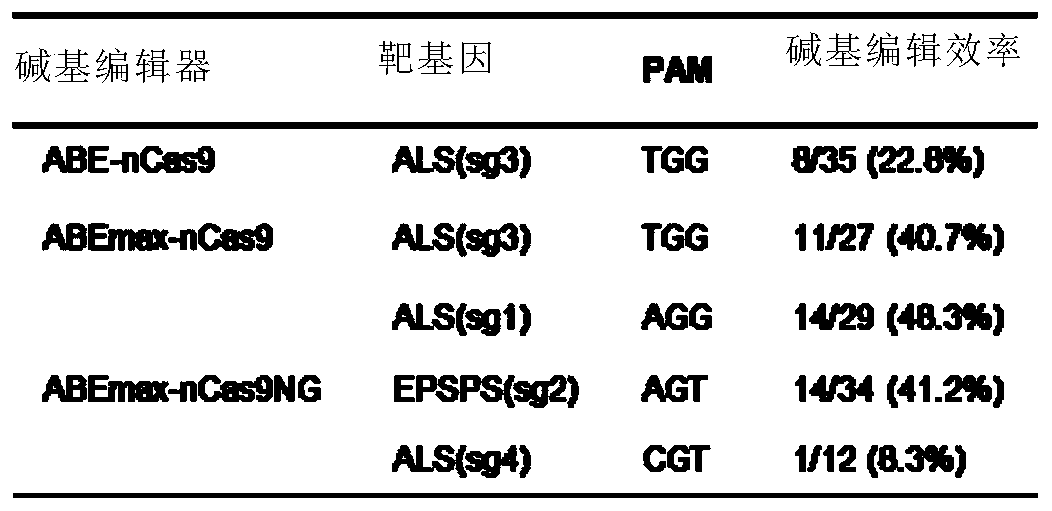
[0244] Example 1 Using ABEmax-nCas9 to improve the base editing efficiency from A to G
[0245] The specific operation process is as follows:
[0246] 1.1 Vector construction
[0247] ABE-nCas9 editor build
[0248] For details, see Hua K, Tao X, Yuan F, Wang D and Zhu J (2018) Precise A.T to G.C Base Editing in the Rice Genome. MOL PLANT11:627-630.
[0249] ABEmax-nCas9 editor build
[0250] (1) Bis-bpNLS, codon-optimized ABE7.10 and bridge sequence composed of 96 bases were synthesized by Nanjing GenScript.
[0251] (2) Replace the ecTadA-linker-ecTadA(7.10)-linker-Cas9-SV40NLS in the original ABE editor with the synthetic sequence using the KpnI and HindIII restriction sites (Hua et al., 2018).
[0252] 1.2 sgRNA design
[0253] Design and synthesize sgRNA for rice ALS gene,
[0254] ALS-sg3: CGCATTCAAGGACATGATCCTGG (SEQ ID NO.: 9)
[0255] ALS-sg1: GCGCCCCCACTTGGGATCATAGG (SEQ ID NO.: 10)
[0256] 1.3 Rice genetic transformation process
[0257] A nucleic acid const...
Embodiment 2
[0268] Example 2 Using ABEmax-nCas9NG to expand the range of base editing
[0269] 1. Carrier construction
[0270] Using Mut The MultiS multiple site-directed mutagenesis kit (Nanjing Nuoweizan) obtained nSpCas9-NG based on nSpCas9(D10A), which contains a total of 7 amino acid substitutions R1335A / L1111R / D1135V / G1218R / E1219F / A1322R / T1337R. The mutated sequence replaced the nSpCas9 (D10A) fragment in the ABEmax-nCas9 editor by BamHI and SpeI restriction sites to obtain the ABEmax-nCas9NG editor.
[0271] In order to verify the editing feasibility of the ABEmax-nCas9NG editor in the non-traditional NGG as the PAM motif, a sgRNA (EPSPS-sg2: GAGAAGGATGCGAAAGAGGAAGT (SEQ ID NO.: 17) and ALS-sg4 was designed in the rice EPSPS and ALS genes respectively. : TAACAAAGAAGAGTGAAGTCCGT (SEQ ID NO.: 18), with AGT and CGT as the PAM motif respectively. Genetic transformation and base editing efficiency detection refer to 1.2 and 1.3 in Example 1. The identification results of transgenic ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com