TOP2A genetic detection probe and preparation method and application thereof
A gene detection and DNA probe technology, applied in the field of TOP2A gene detection probe and its preparation, can solve the problems of long hybridization time, low hybridization efficiency, and high signal background
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
preparation example Construction
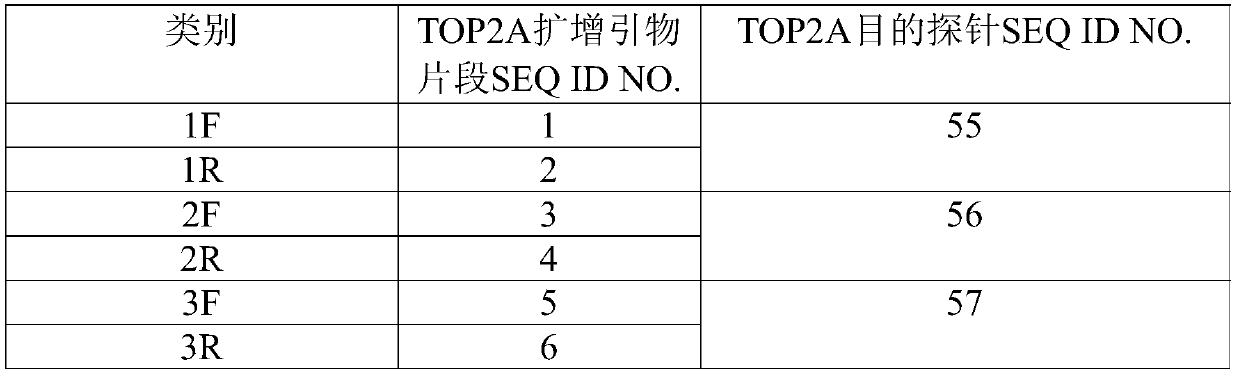
[0040] The invention provides a method for preparing a TOP2A gene detection probe. The prepared TOP2A gene detection probe includes a TOP2A single-strand probe for the TOP2A gene region and a control single-strand probe for the centromere region of chromosome 17.
[0041] One embodiment of the present invention provides a method for preparing a TOP2A single-stranded probe, comprising the steps of:
[0042] Step 1: Select the TOP2A gene region as the TOP2A target gene, design TOP2A single-stranded candidate probes according to the TOP2A target gene, remove the TOP2A single-stranded candidate probes that non-specifically hybridize and overlap with the TOP2A target gene to obtain the TOP2A target probe, and obtain the TOP2A target probe by screening Design TOP2A amplification primer fragments upstream and downstream of the TOP2A target probe.
[0043] In the above step 1, select the TOP2A gene region as the TOP2A target gene, and design the TOP2A single-stranded candidate probes ...
Embodiment 1
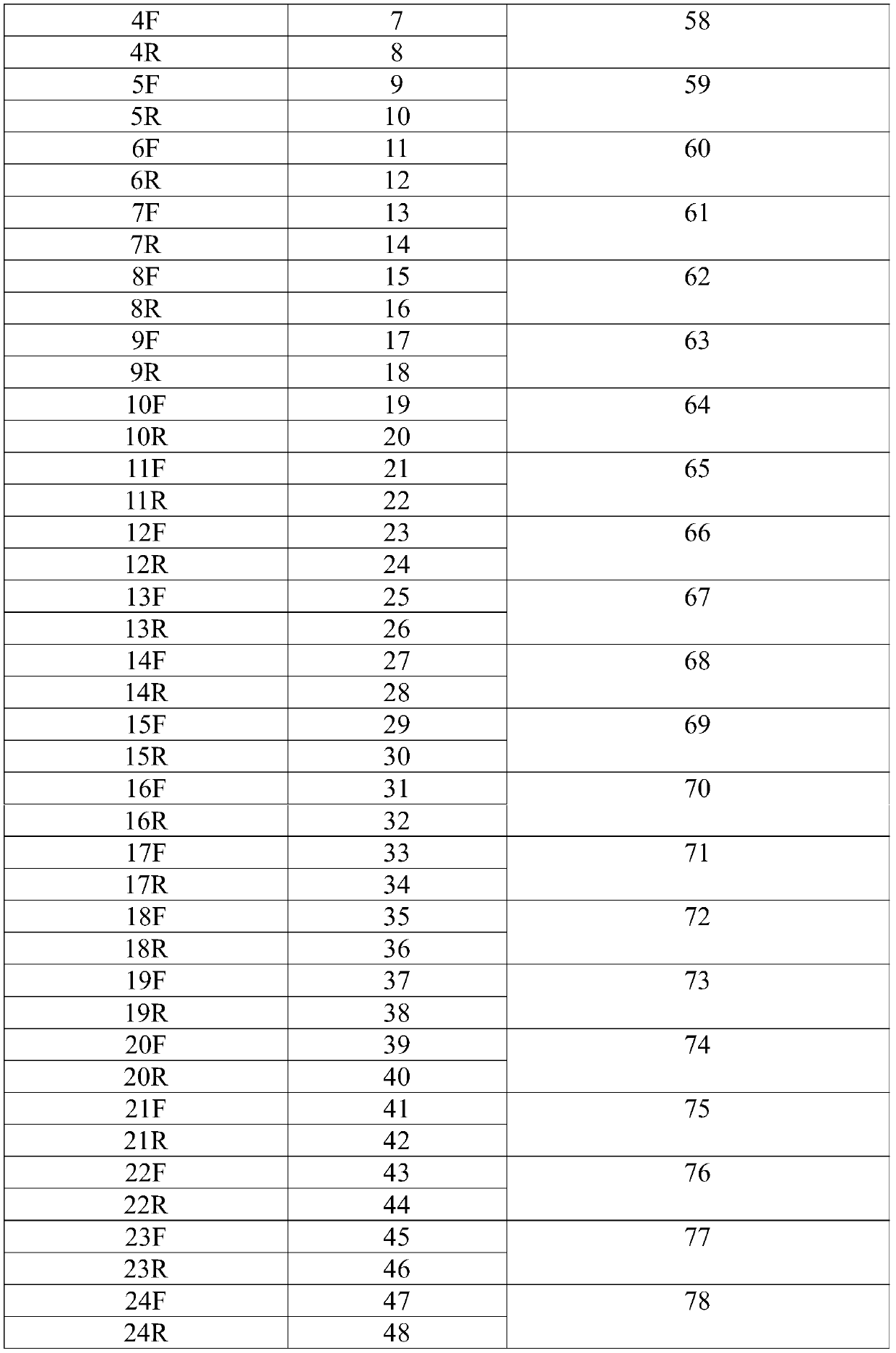
[0082] Select the TOP2A gene region as the TOP2A target gene, design a TOP2A single-stranded candidate probe every 3bp, the length of each TOP2A single-stranded candidate probe is 200nt, and design 40 TOP2A single-stranded candidate probes; through BLAST comparison, Remove TOP2A single-stranded candidate probes located in the repetitive region of the TOP2A gene, remove TOP2A single-stranded candidate probes with non-specific Tm values between 75-85°C by scoring, and select non-overlapping TOP2A single-stranded candidate probes as TOP2A targets As for the probes, a total of 26 TOP2A target probes were obtained, and the sequences of the 26 TOP2A target probes are shown in SEQ ID NO.55-80. The TOP2A amplification primer fragments were designed for the upstream and downstream of the 26 TOP2A target probes screened. The sequences of TOP2A amplification primer fragments are shown in SEQ ID NO.1-52. The TOP2A amplification primer fragment and the corresponding TOP2A target probe a...
Embodiment 2
[0120] Select the centromere region of chromosome 17 as the control gene, design a single-stranded fragment with a length of 206 nt as the control target probe for the control gene, and design control amplification primer fragments for the upstream and downstream of the control target probe.
[0121] The sequences of the control amplification primer fragments are shown in SEQ ID NO.53-54, wherein the sequence shown in SEQ ID NO.53 is the upstream amplification primer, and the sequence shown in SEQ ID NO.54 is the downstream amplification primer. The sequence of the control target probe is shown in SEQ ID NO.81.
[0122] PCR is used to amplify the control target probe to obtain a PCR product.
[0123] To set up a PCR reaction:
[0124]
[0125] Purification of PCR products: Combine the PCR products, add 1ml of butanol, and mix well; then add 400μl Orange-DX, mix well; centrifuge for 2min, remove the upper organic phase, and pass the aqueous phase through the column to bind. ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com