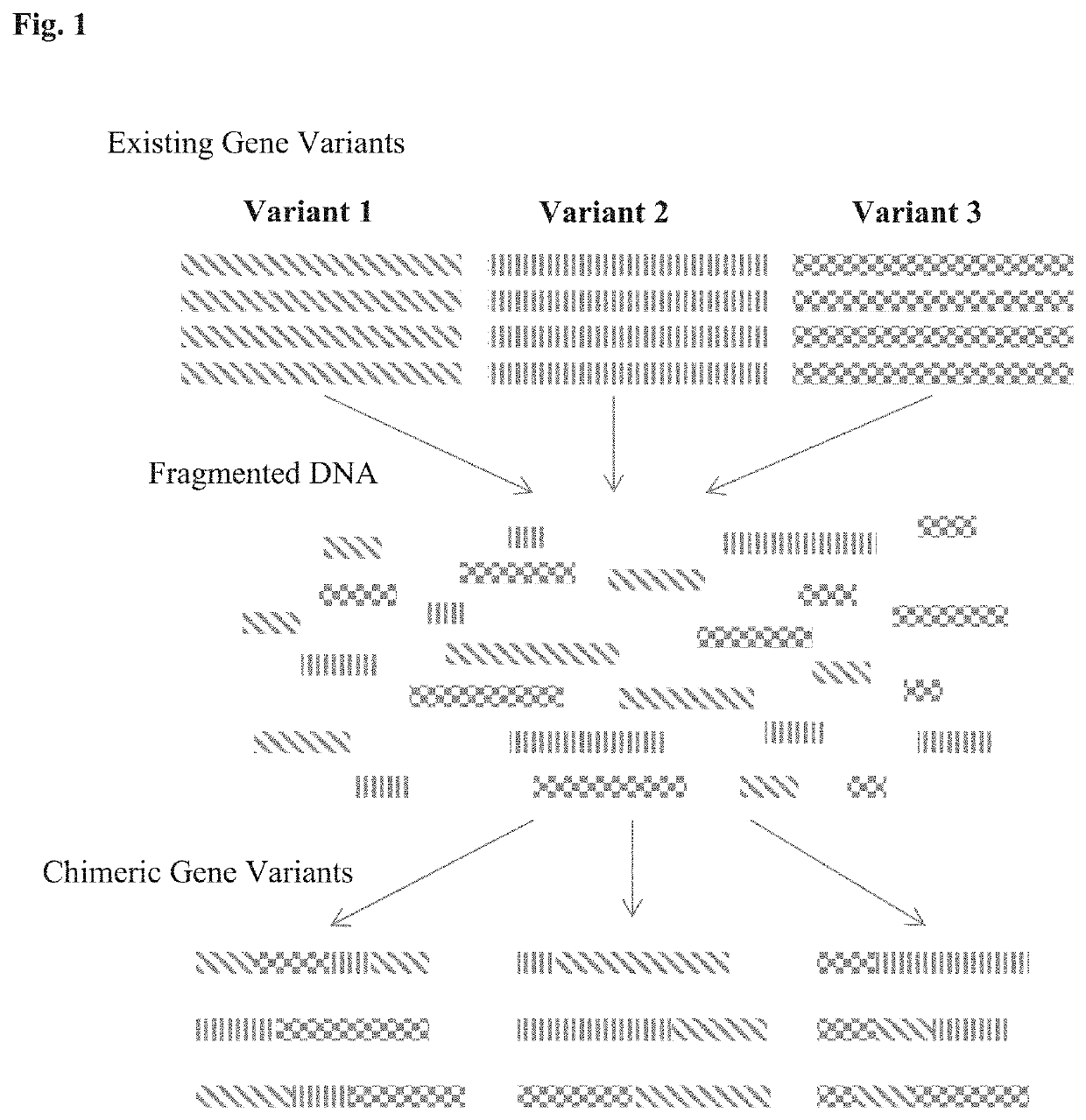
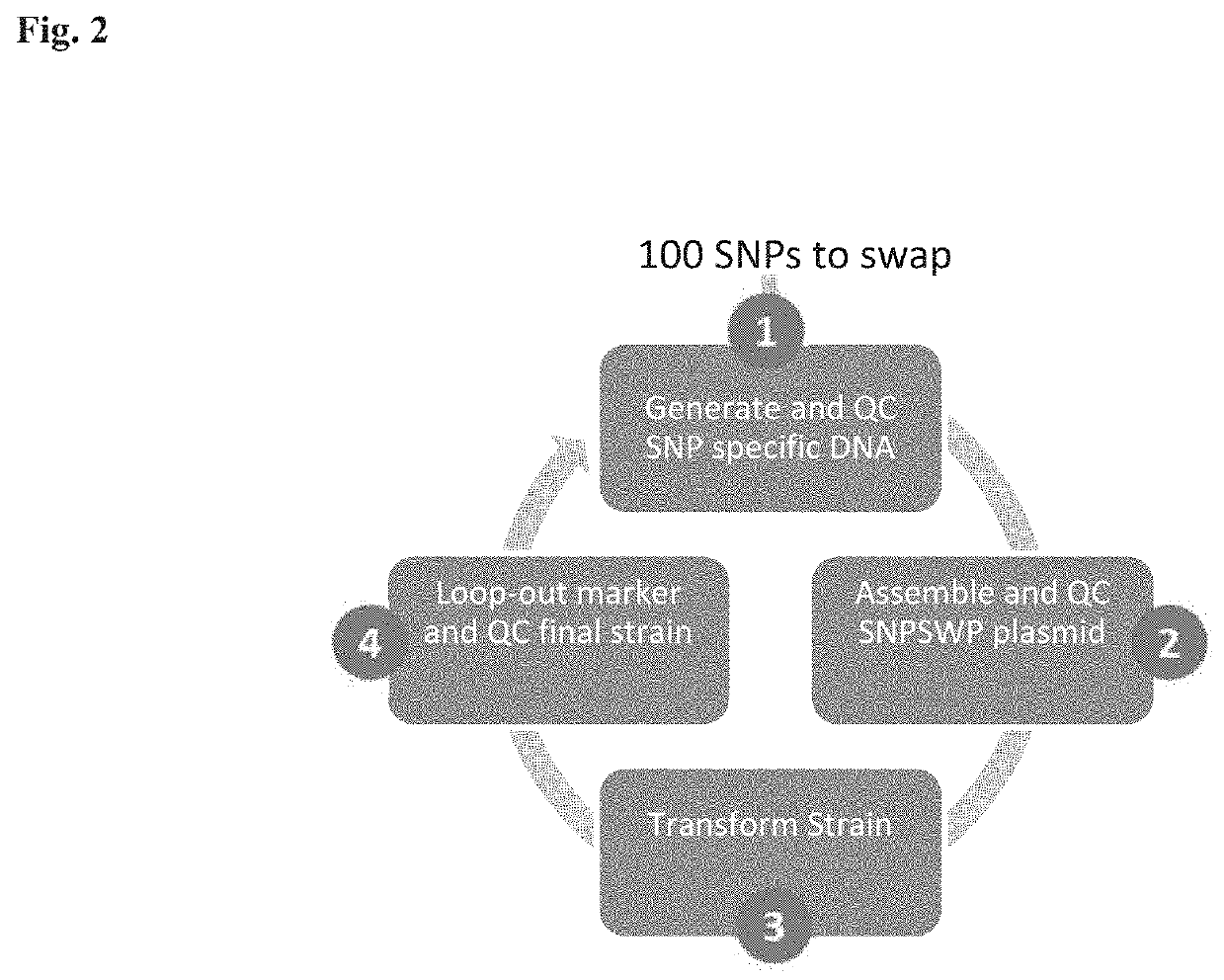
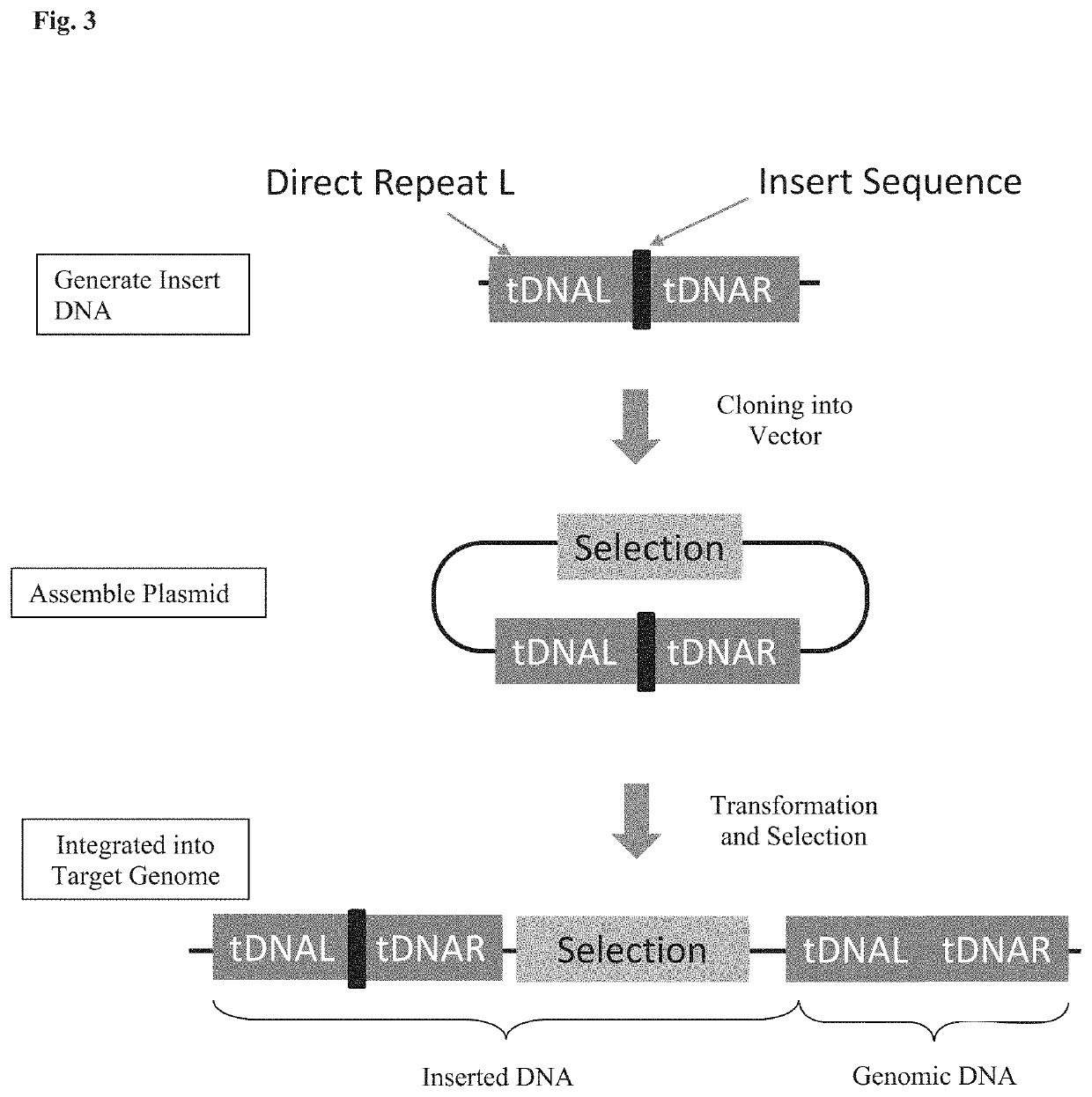
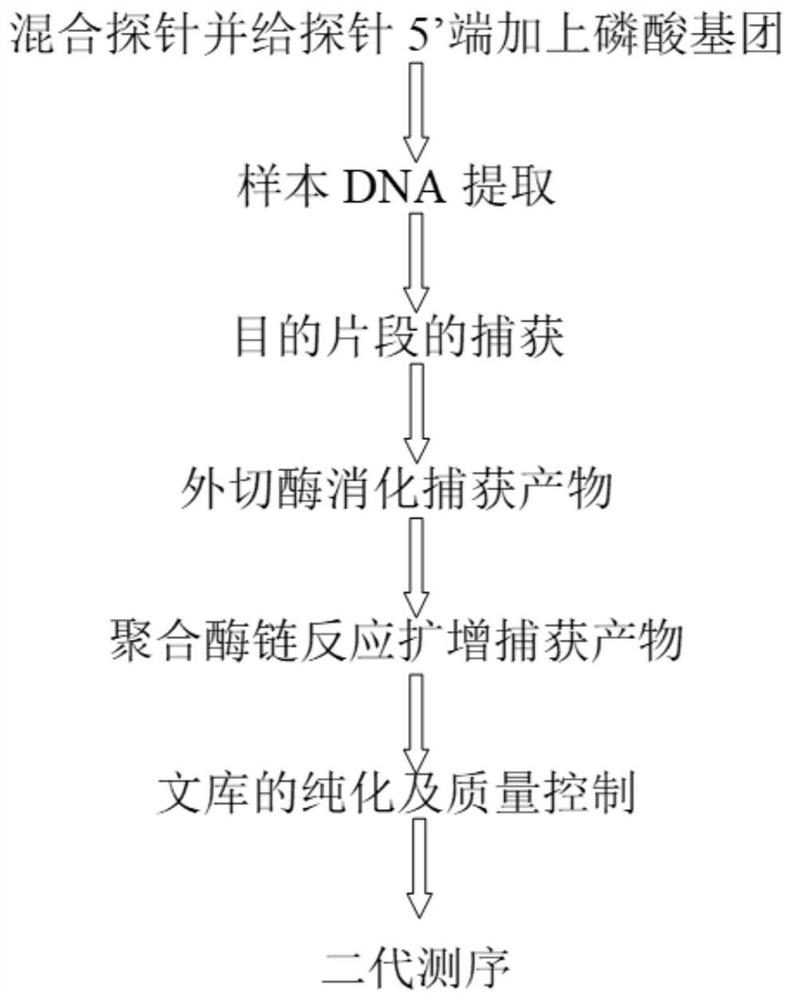
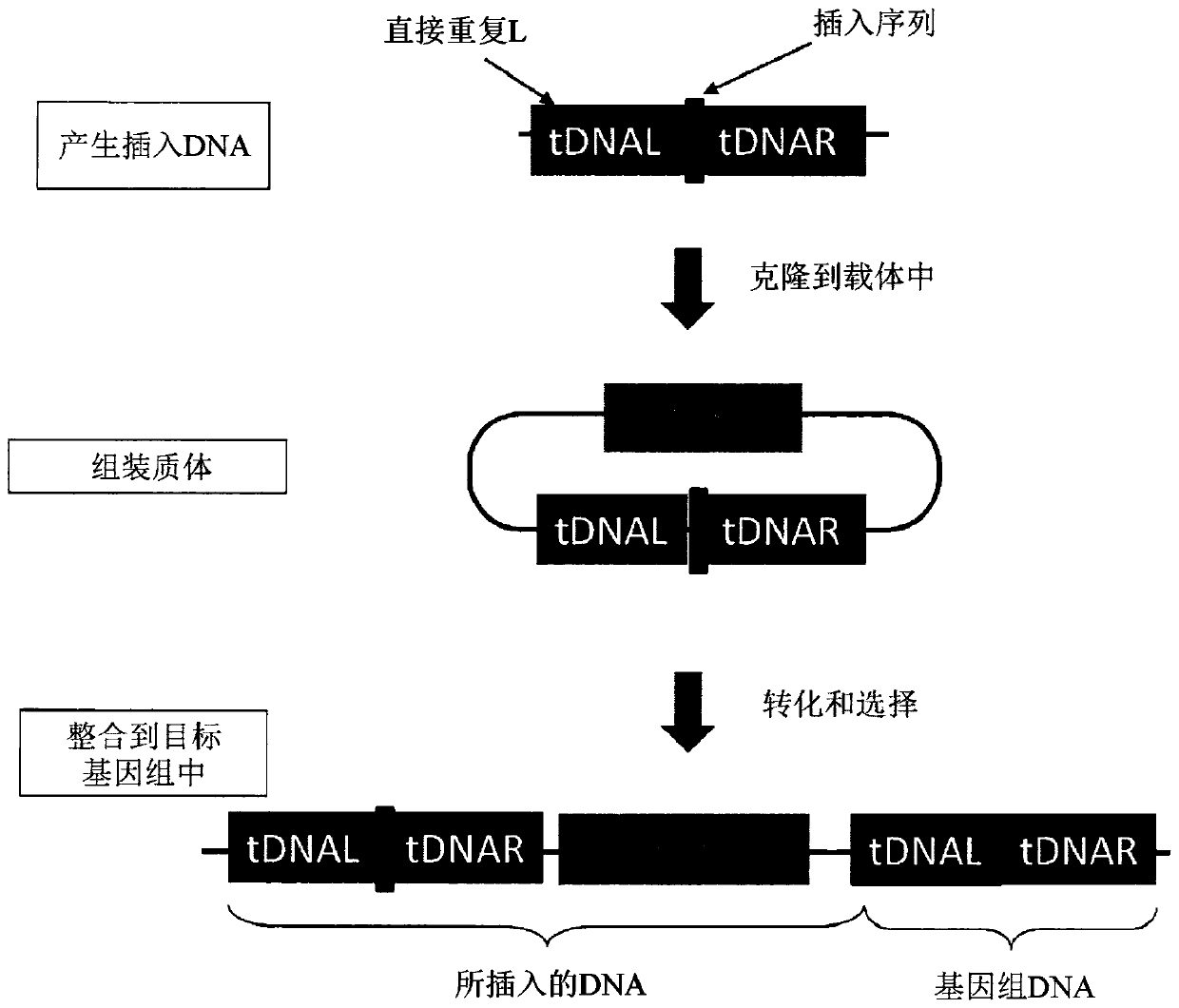
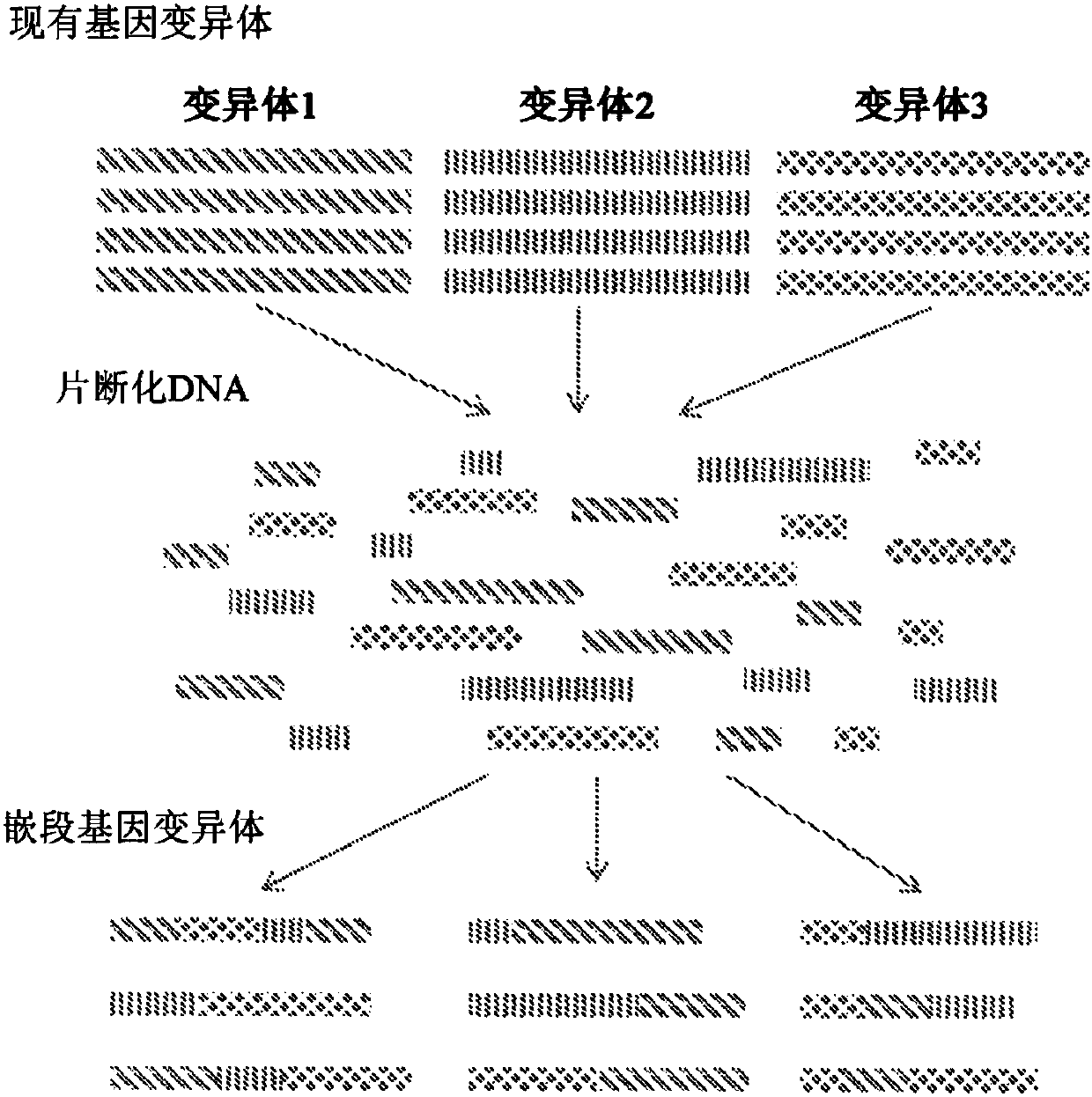
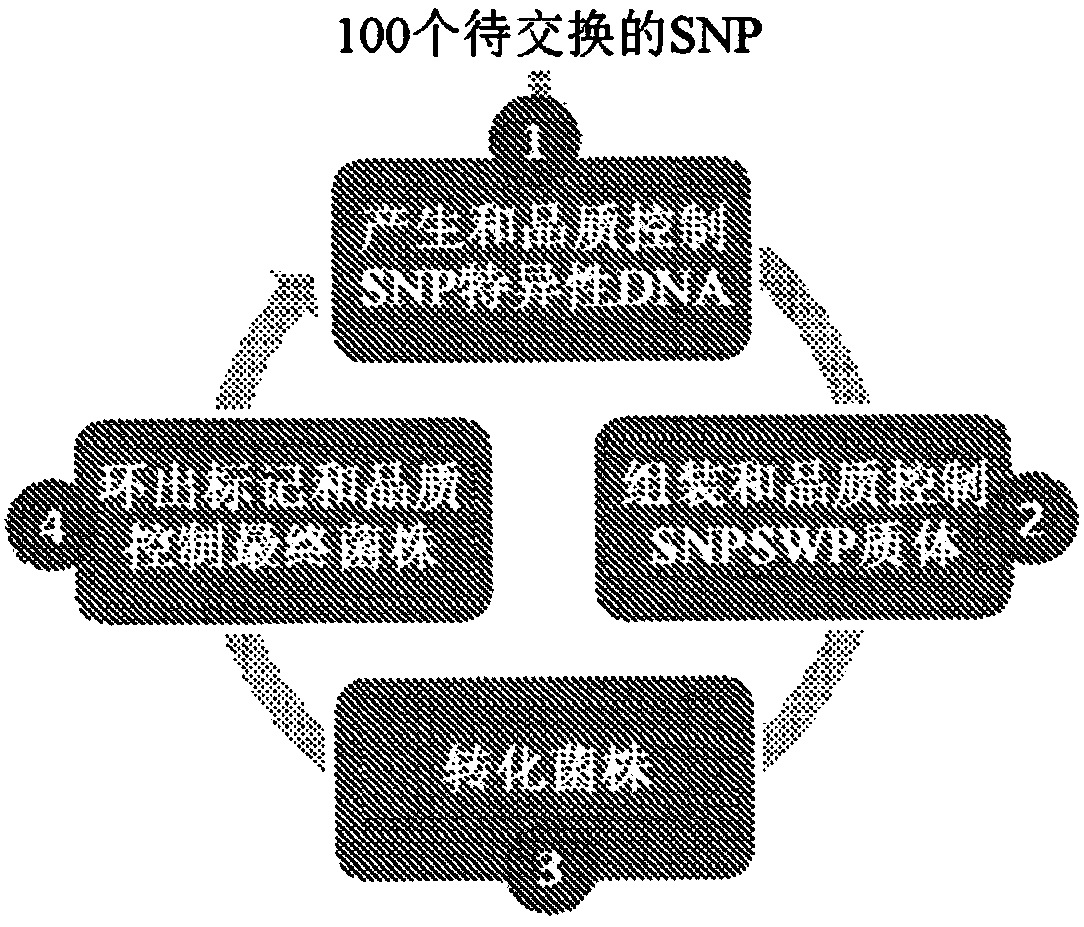
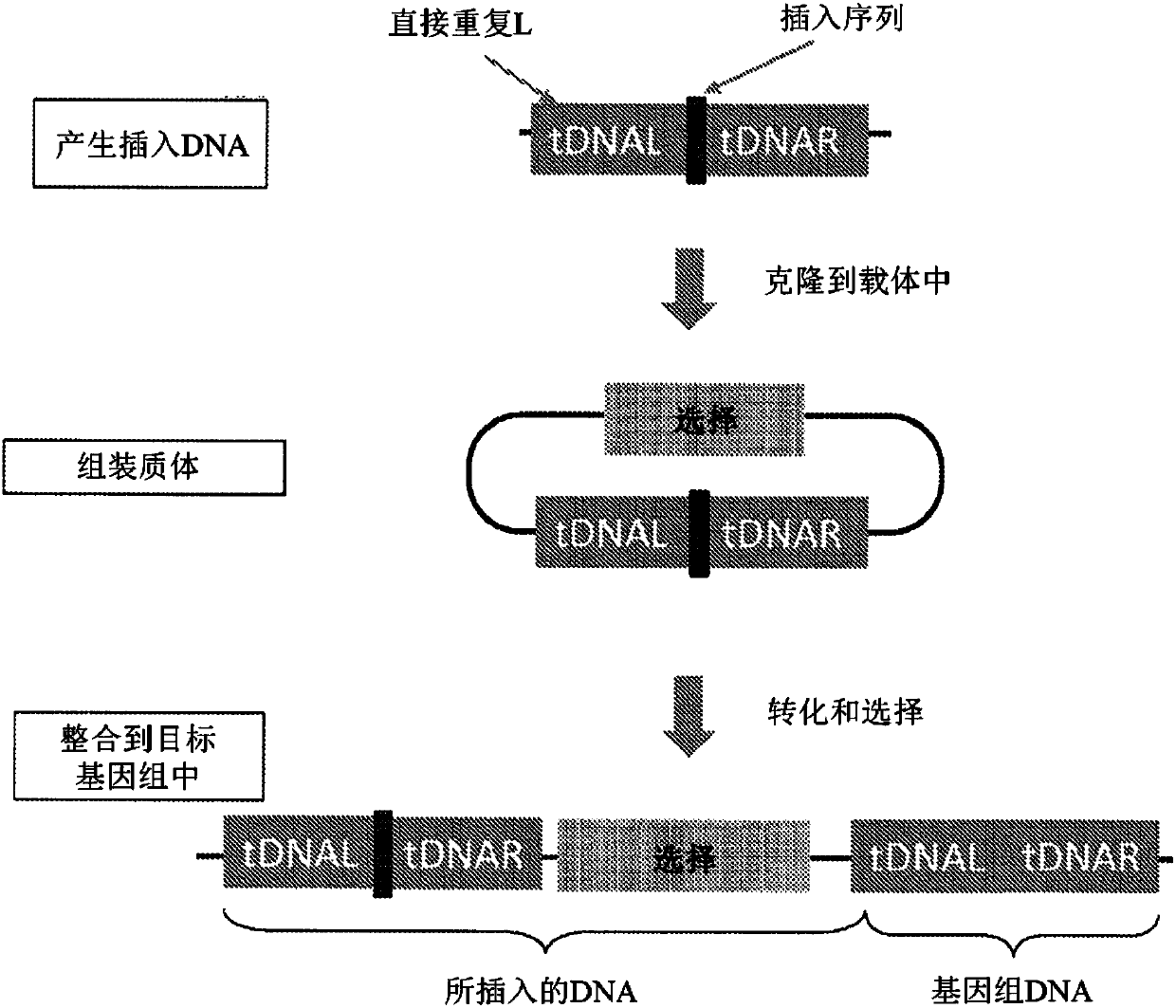
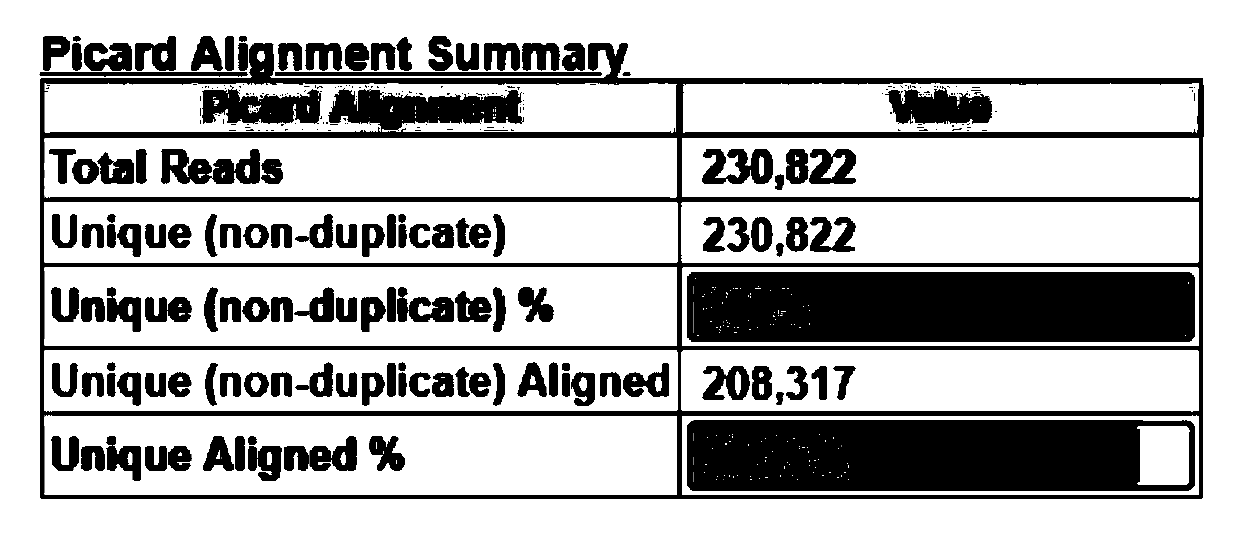
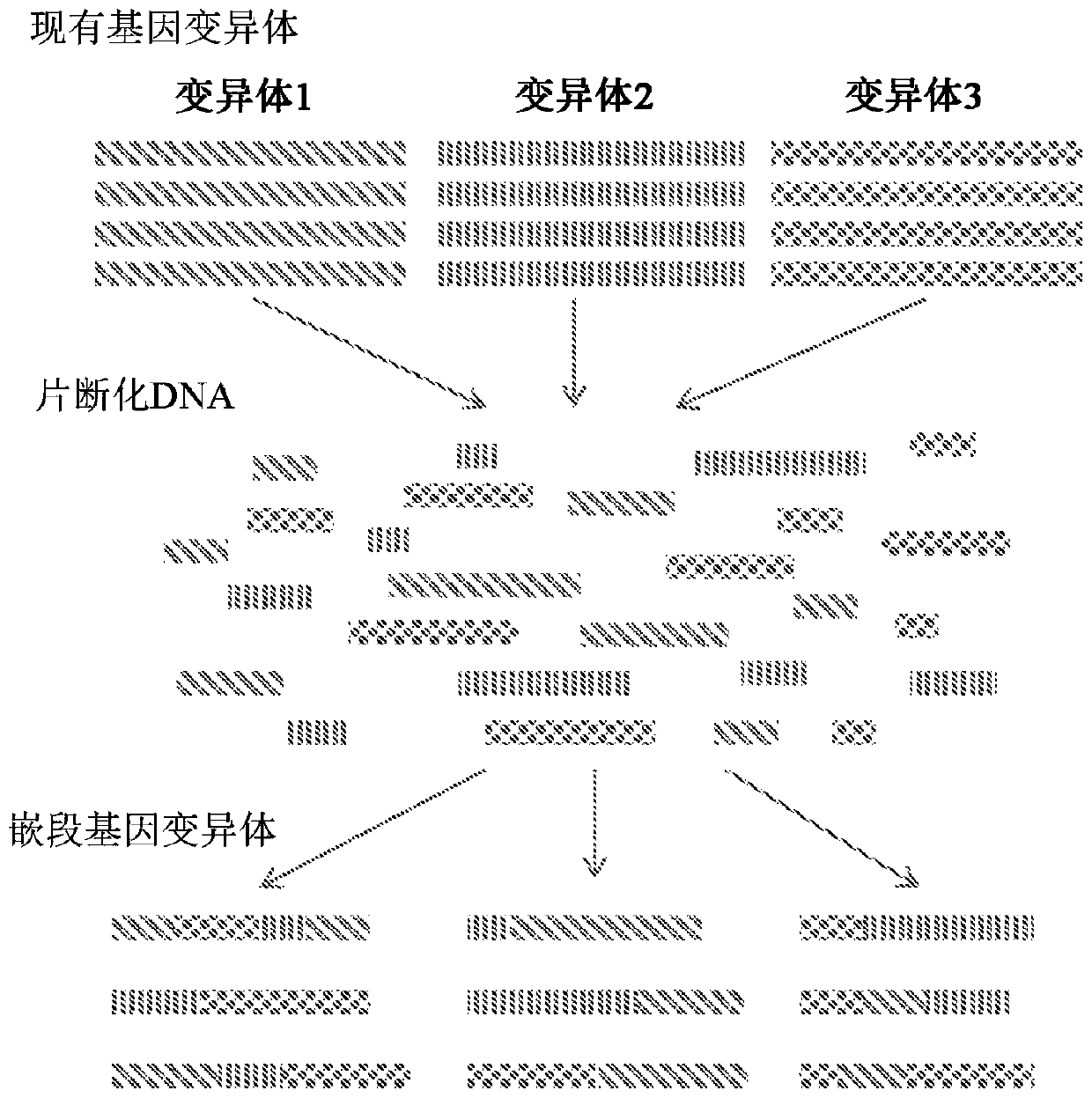
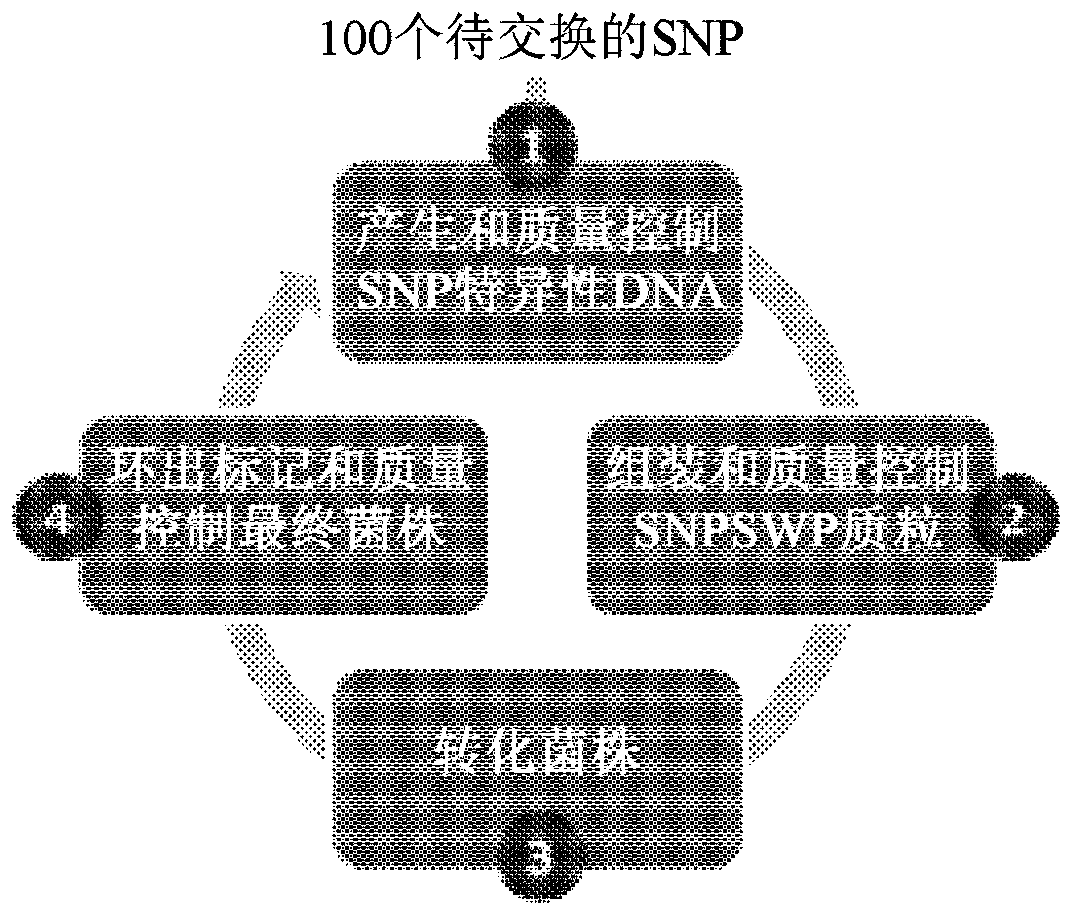
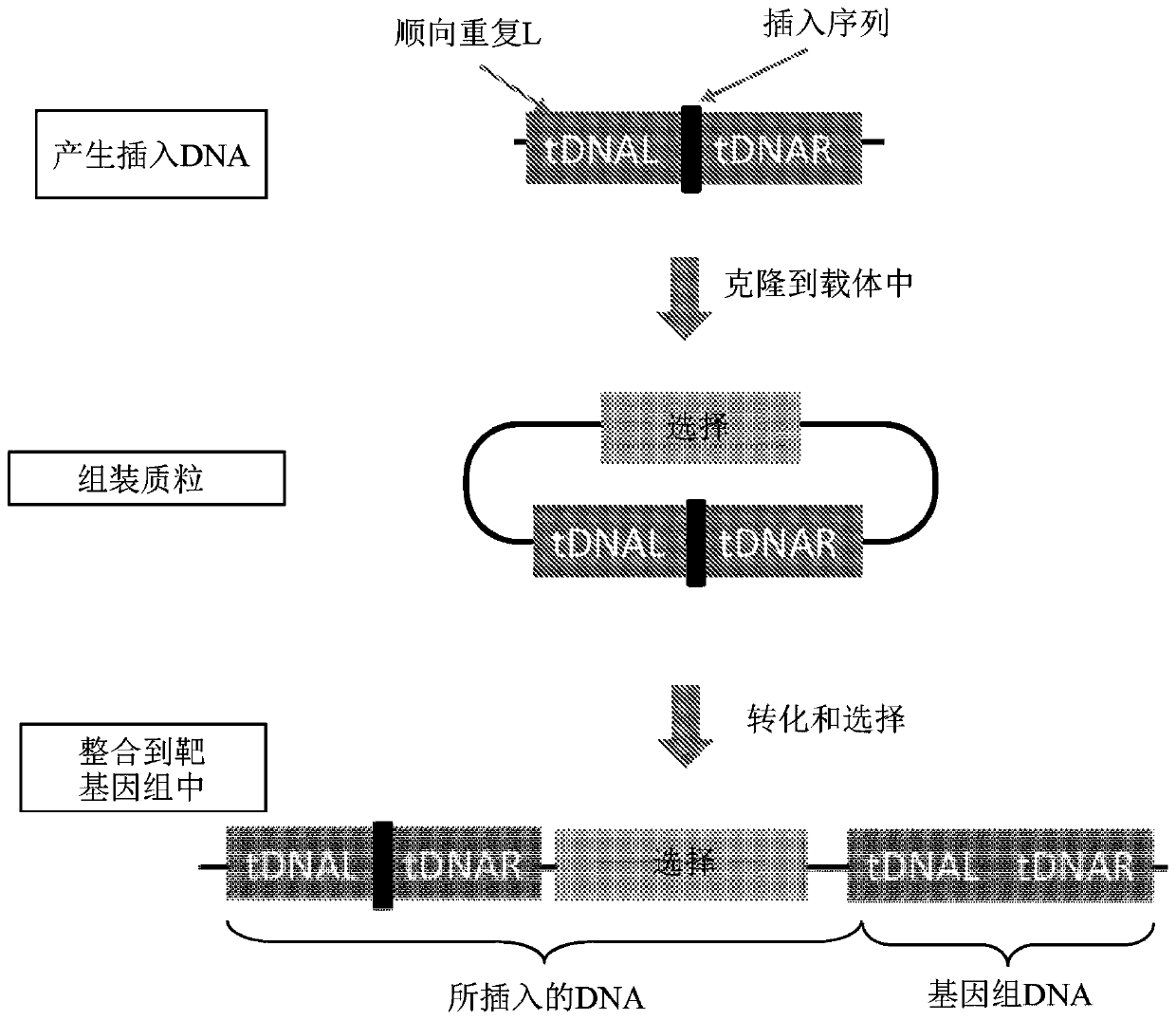
Patents
Literature
51 results about "Genetic design" patented technology
Efficacy Topic
Property
Owner
Technical Advancement
Application Domain
Technology Topic
Technology Field Word
Patent Country/Region
Patent Type
Patent Status
Application Year
Inventor
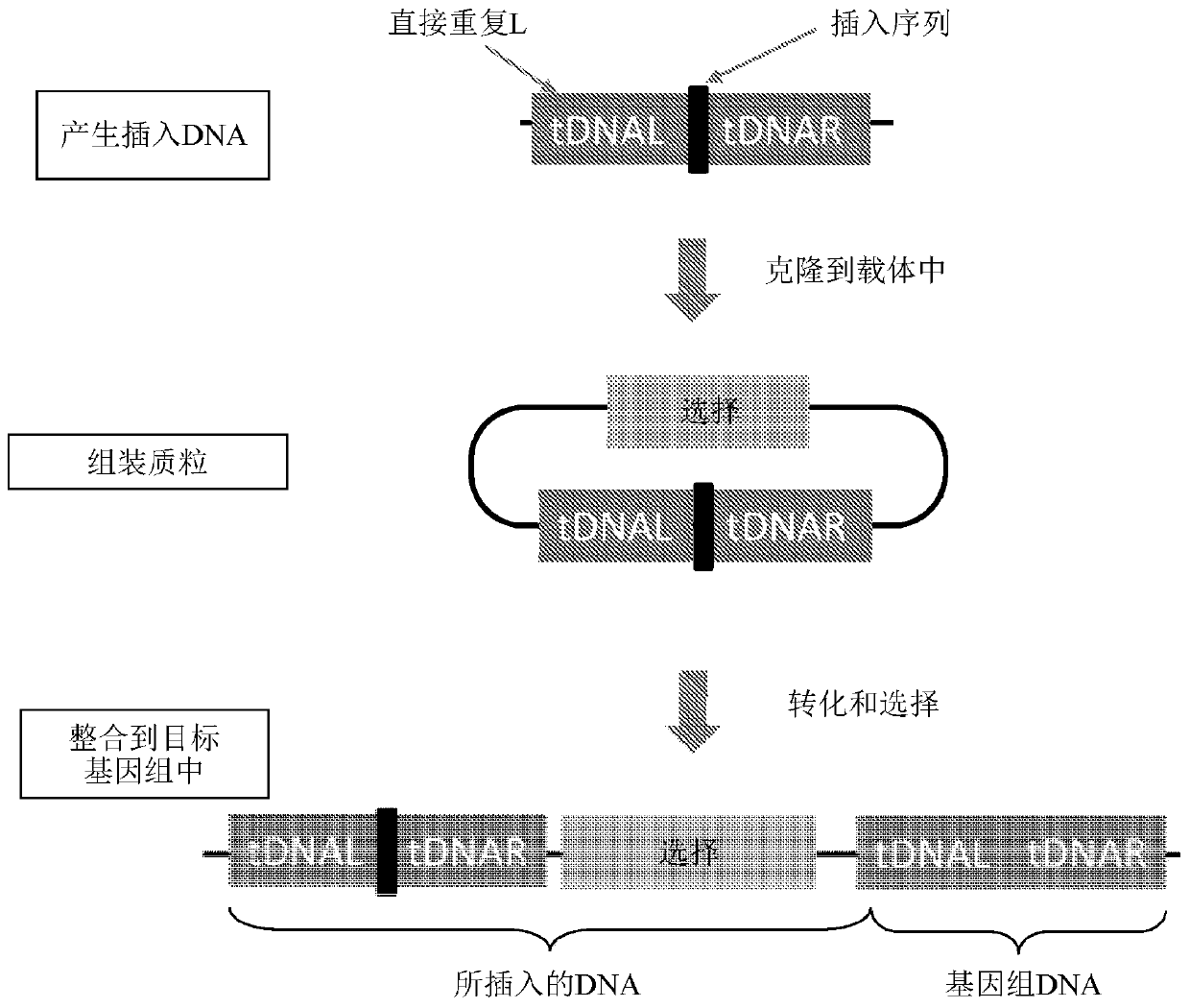
Principle of quantitative trait locus (QTL) positioning
The invention relates to a principle of quantitative trait locus (QTL) positioning. The QTL refers to the position of a gene for controlling quantitative traits in a genome; A polygenic system for controlling the quantitative trait is used as integration for research on the basis of classical quantitative genetics of polygene hypothesis by several genetic designs and statistical models; and the genetic characteristics of the quantitative traits are described by genetic parameters, such as hereditary capacity and genetic variance.
Owner:李祥
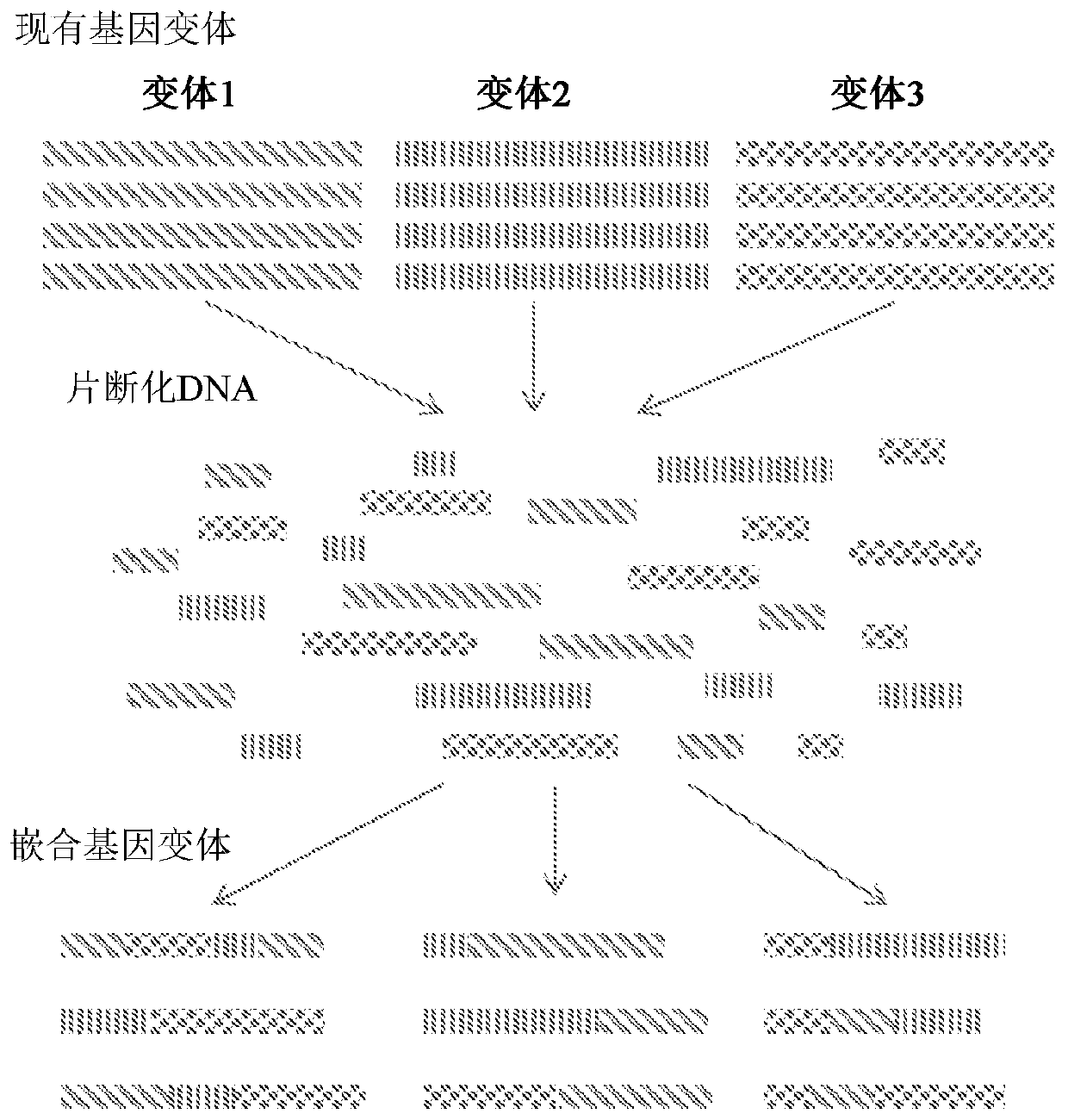
Amplification internal standard preparation based on DNA stochastic shuffling technology
ActiveCN101503735AImprove accuracyEnsure consistencyMicrobiological testing/measurementMicroorganism based processesSoftware EvaluationPathogenic microorganism
The invention provides a method for preparing a proliferation internal label based on a DNA random reorganization technique, and belongs to the field of the biological technology. The method comprises the following steps that: according to a target gene design, a specific detection primer and a TaqMan probe are obtained; according to DNA sequences on the probe combination part of a target fragment, DNA random reorganization software randomly generates reorganization sequences, and the generated reorganization sequences replace the DNA sequences on the probe combination part of the target fragment respectively to generate corresponding proliferation internal label sequences to be screened; by comparing the fluorescent probe design software with the BLAST-N software, the proliferation internal label sequences are screened to obtain the sequence which has the highest software evaluation value and is not homologous to the sequences of other pathogenic microorganism genomes, and the sequence is used as the proliferation internal label sequence; the proliferation internal label sequence obtained from the third step is cloned to a vector to construct the vector containing the proliferation internal label sequence; and the obtained proliferation internal label sequence is detected. The method can help to display the existence of the inhibition phenomenon in detection, thereby improving the accuracy rate of the detection result.
Owner:SHANGHAI JIAO TONG UNIV
Primer probe composition, kit and method for detecting coronavirus 2019-nCoV
InactiveCN111088405ARealize detectionHigh sensitivityMicrobiological testing/measurementAgainst vector-borne diseasesVirusGene
The invention relates to a primer probe composition, kit and method for detecting new coronavirus 2019-nCoV. A composition one and a composition two are designed as the primer probe composition, a specific primer probe is designed for multiple target genes of the virus based on the digital PCR technology, the digital PCR detection can directly count a number of positive micro-reaction systems of the PCR terminal point, Poisson statistics is used to directly calculate an absolute copy number of a target molecule, the detection of a single target molecule can be realized, the absolute count andquantitative detection of the virus content is achieved, and in addition, the entire detection time does not exceed 3 h, so that the method is time-saving and efficient, and has high sensitivity.
Owner:苏州行知康众生物科技有限公司
HTP genomic engineering platform for improving escherichia coli
PendingCN110945125AVector-based foreign material introductionDNA preparationEscherichia coliGenomic engineering
The present disclosure provides a HTP genomic engineering platform for improving escherichia coli. that is computationally driven and integrates molecular biology, automation, and advanced machine learning protocols. The integrative platform utilizes a suite of HTP molecular tool sets to create HTP genetic design libraries, which are derived from, inter alia, scientific insight and iterative pattern recognition.
Owner:ZYMERGEN INC
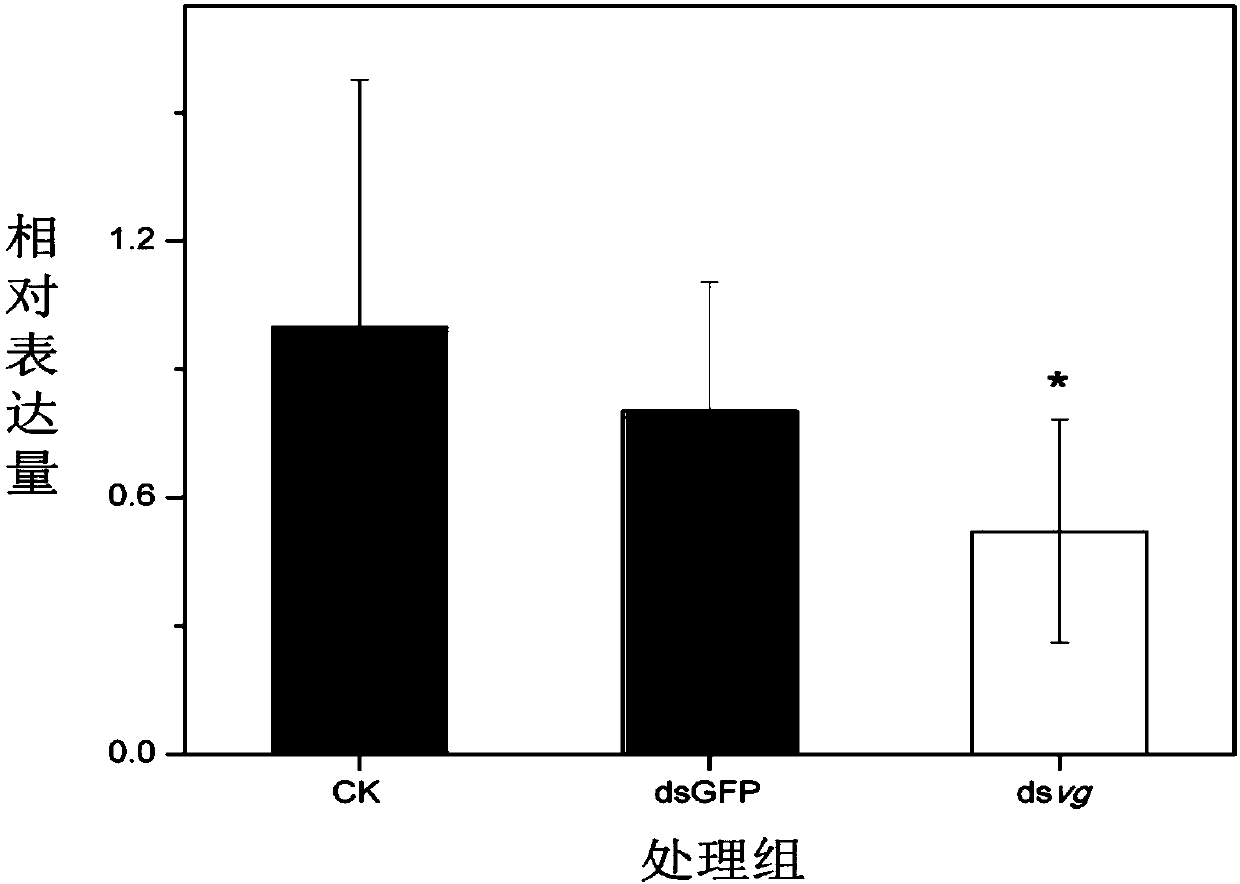
dsRNA of wing development related gene vestigial and application thereof in control of citrus fruit fly
ActiveCN107916264AEasy to operateGuaranteed Interference EfficiencyBiocideAnimal repellantsSocial benefitsHuman health
The invention discloses dsRNA of wing development related gene vestigial and application thereof in control of citrus fruit fly. A primer is designed for citrus fruit fly development related genes anddsRNA is synthesized first, and application of RNAi is performed on the citrus fruit fly development related genes by adopting a feeding method, so that phenotype of wing developmental malformation is obtained on the premise of ensuring interference efficiency. The dsRNA of wing development related gene vestigial and application thereof in control of citrus fruit fly disclosed by the invention solve the problem that injection implantation of dsRNA is difficul to operate in the field, lay a foundation for future study of directly sprayed RNA preparations to control the citrus fruit fly, and have excellent economic and social benefits for effectively controlling invasion and diffusion of the citrus fruit fly and protecting agricultural production and ecology security and human health in China.
Owner:CHINA AGRI UNIV
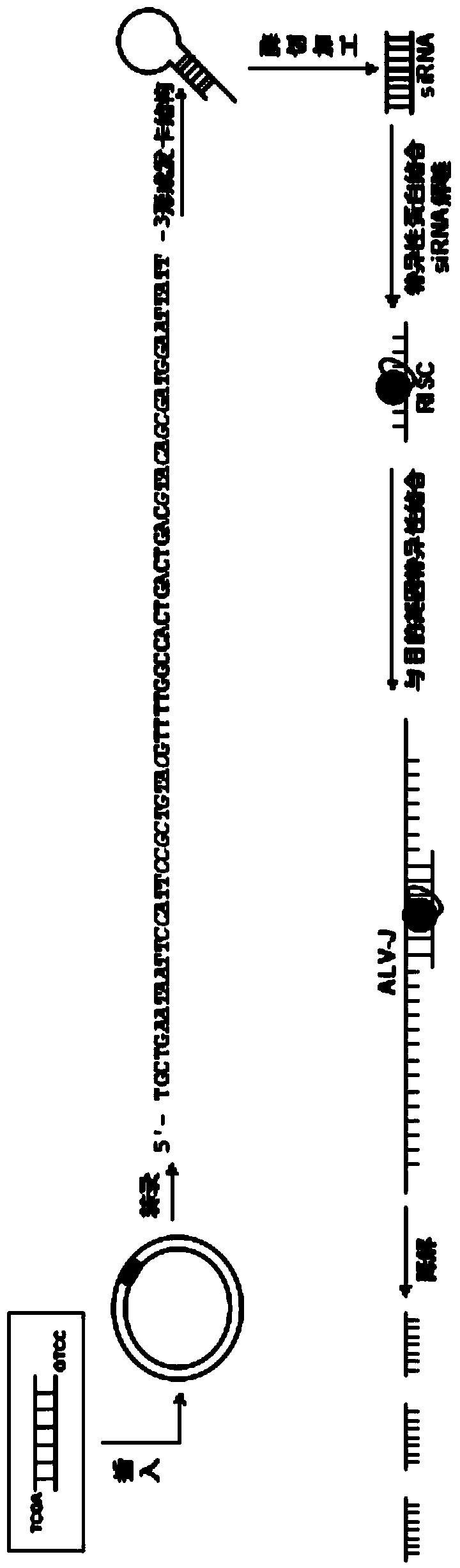
J avian leukosis virus subgroup env gene conserved sequence-based siRNA (Small Interfering RNA (Ribonucleic Acid)) recombinant interference carrier as well as preparation method and application thereof
InactiveCN103805635AEffective interferenceReduce economic lossGenetic material ingredientsAntiviralsConserved sequenceAvian leukosis viruses
The invention relates to the technical field of genetic engineering, and provides a J avian leukosis virus subgroup env gene conserved sequence-based siRNA (Small Interfering RNA (Ribonucleic Acid)) recombinant interference carrier. The preparation method comprises the following steps: on the basis that the env gene has important meaning for ALV (avian leukosis virus), designing and synthesizing siRNA by the env gene, building to form a hairpin structure of siRNA, further obtaining annealing double-stranded DNA (Deoxyribonucleic Acid), and connecting the double-stranded DNA with the carrier to build the recombinant interference carrier. After the interference carrier and the virus co-transfect cells are adopted, the interference carrier provided by the invention can effectively interfere the transcription and the duplication of the J avian leukosis virus subgroup in the in-vitro cell and the live chicken, the scientific base and the technical support can be provided for preventing and curing the J avian leukosis virus subgroup, and the economic loss caused by the ALV-J infection in the poultry industry can be reduced.
Owner:SHANDONG AGRICULTURAL UNIVERSITY
A htp engineering platform
ActiveUS20210261949A1Improving phenotypic performanceHeating or cooling apparatusBiostatisticsMicroorganismBiochemical engineering
The present disclosure provides a HTP microbial genomic engineering platform that is computationally driven and integrates molecular biology, automation, and advanced machine learning protocols. This integrative platform utilizes a suite of HTP molecular tool sets to create HTP genetic design libraries, which are derived from, inter alia, scientific insight and iterative pattern recognition. The HTP genomic engineering platform described herein is microbial strain host agnostic and therefore can be implemented across taxa. Furthermore, the disclosed platform can be implemented to modulate or improve any microbial host parameter of interest.
Owner:ZYMERGEN INC
High-throughput targeted library building method and application
PendingCN112266948AReduce biasEasy to operateMicrobiological testing/measurementLibrary creationMultiplexBase J
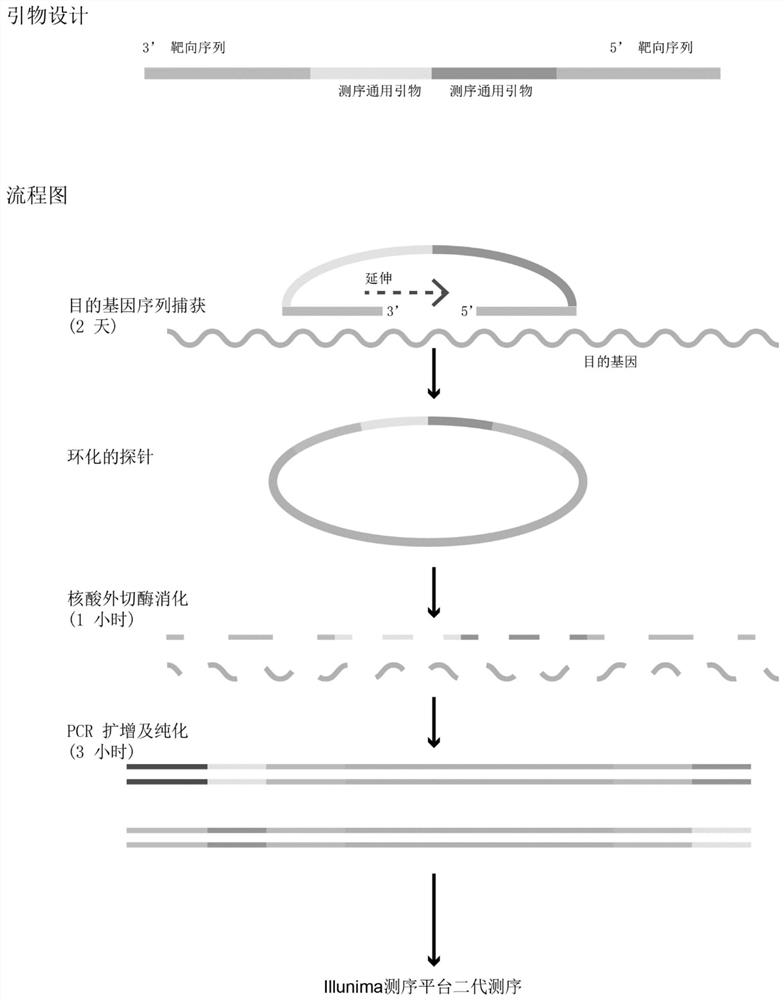
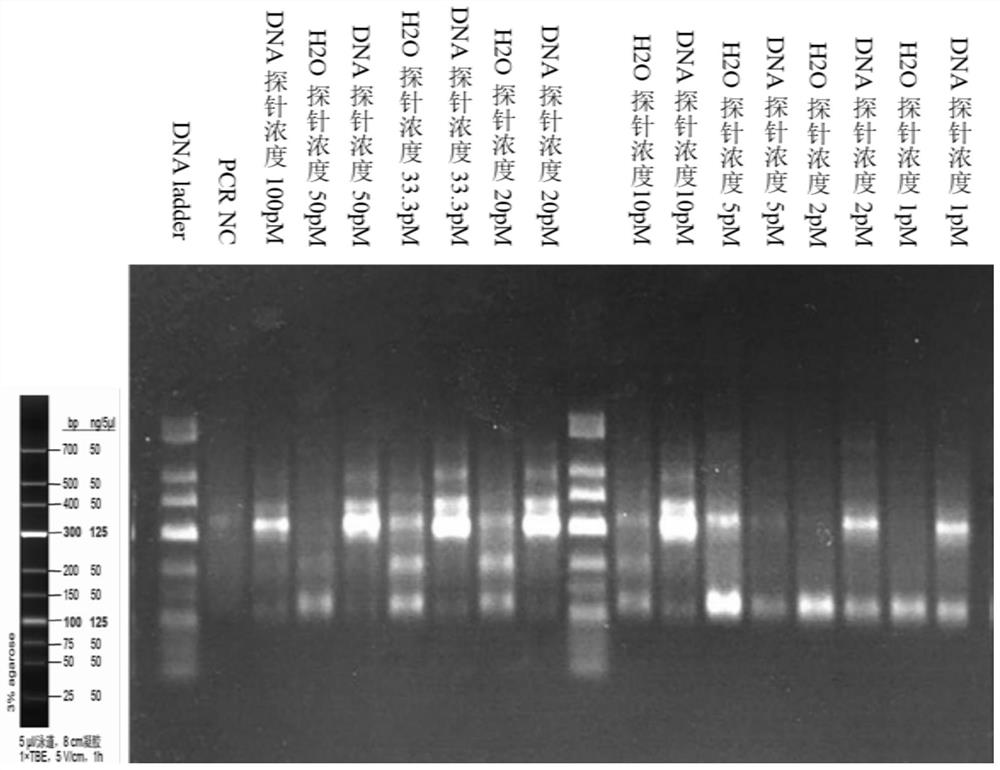
The invention discloses a high-throughput targeted library building method and application. The high-throughput targeted library building method comprises the following steps that a targeted capture probe and a PCR amplification primer are designed and synthesized based on a detected gene; a genome DNA of a sample is hybridized with the targeted capture probe overnight to obtain a captured targetfragment, and cyclizing, digesting, purifying and PCR amplification are carried out to obtain a constructed targeted sequencing library; and the targeted sequencing library is purified, the concentration and size of DNA fragments of the library are detected, second-generation sequencing is carried out, and the quality and sequencing information of the targeted sequencing library are analyzed according to a sequencing result. According to the high-throughput targeted library building method and application, hundreds of detection genes can be amplified in a round of PCR reaction, so that the defects that only a small number of gene loci can be detected by multiplex PCR library construction and experimental conditions are difficult to optimize are overcome; only one turn of PCR is performed,and random barcode are arranged on a probe framework, so that PCR bias is reduced, and base errors introduced by PCR and sequencing can be distinguished; and the library building step is simplified, and the reagent and time cost is reduced.
Owner:SUN YAT SEN MEMORIAL HOSPITAL SUN YAT SEN UNIV
Accurate method capable of carrying out in-vivo target activity evaluation and off-target detection in batches
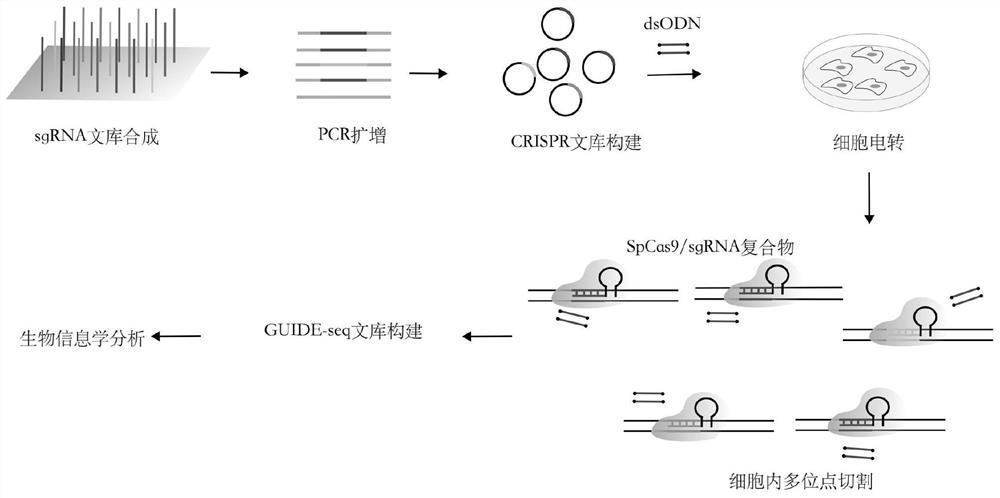
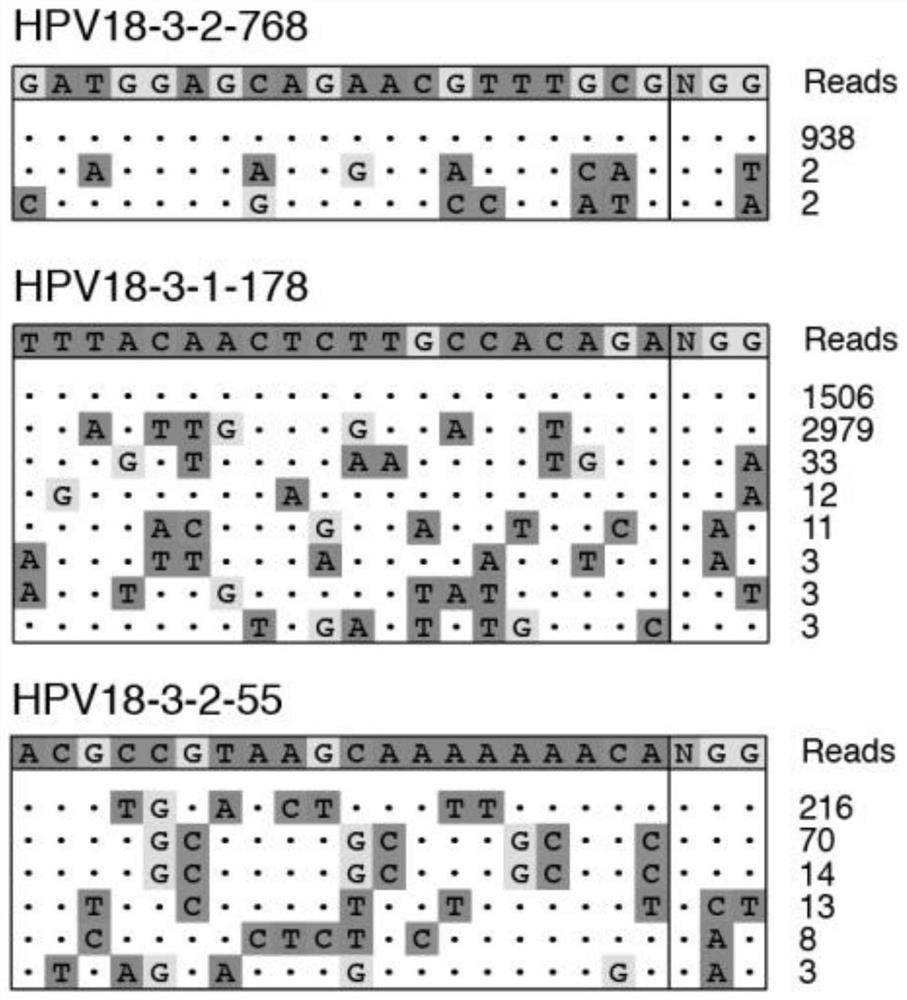
ActiveCN112501252AImprove throughputHigh detection throughputMicrobiological testing/measurementLibrary creationIntracellularPlasmid
Owner:ZHUHAI SHU TONG MEDICAL TECH CO LTD
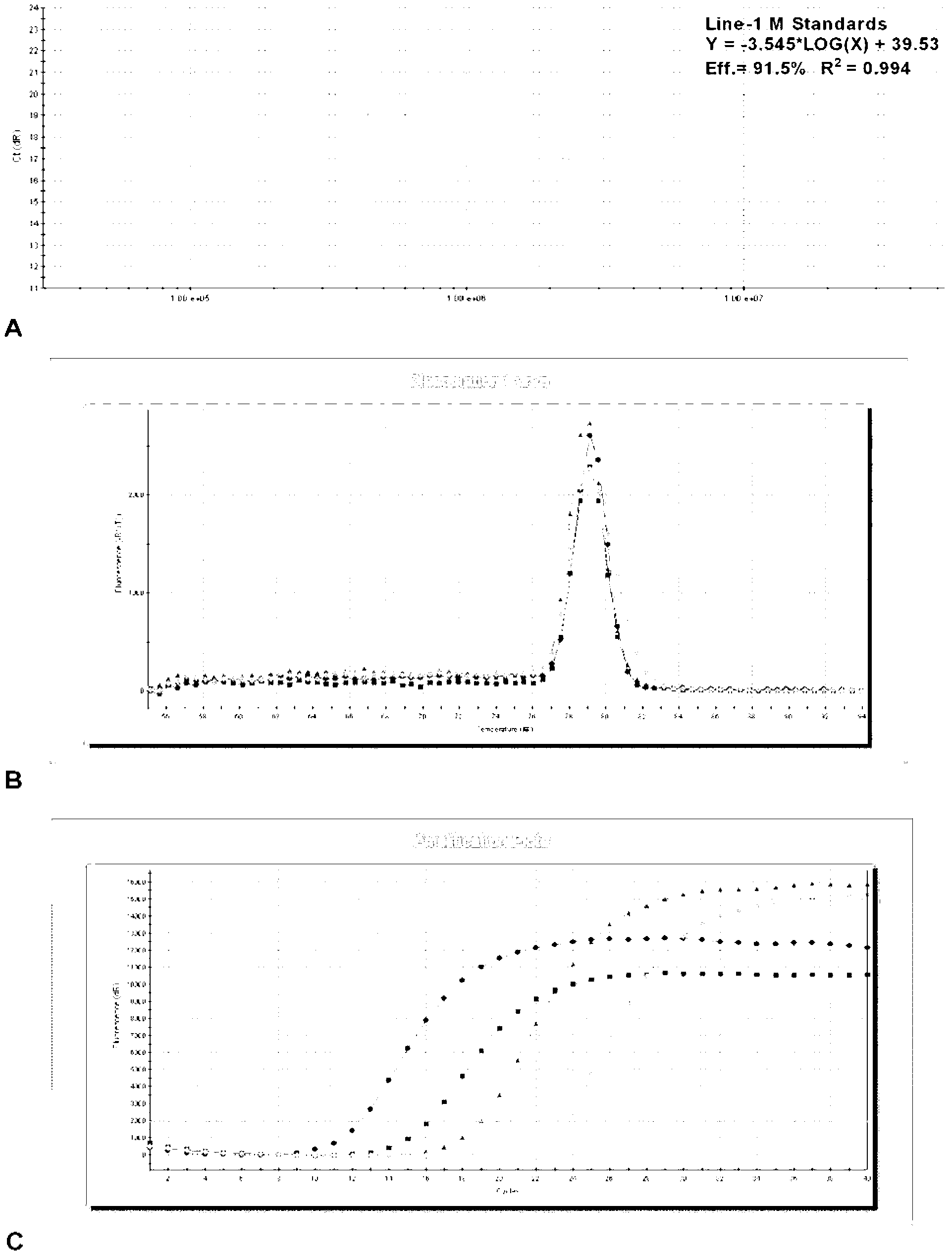
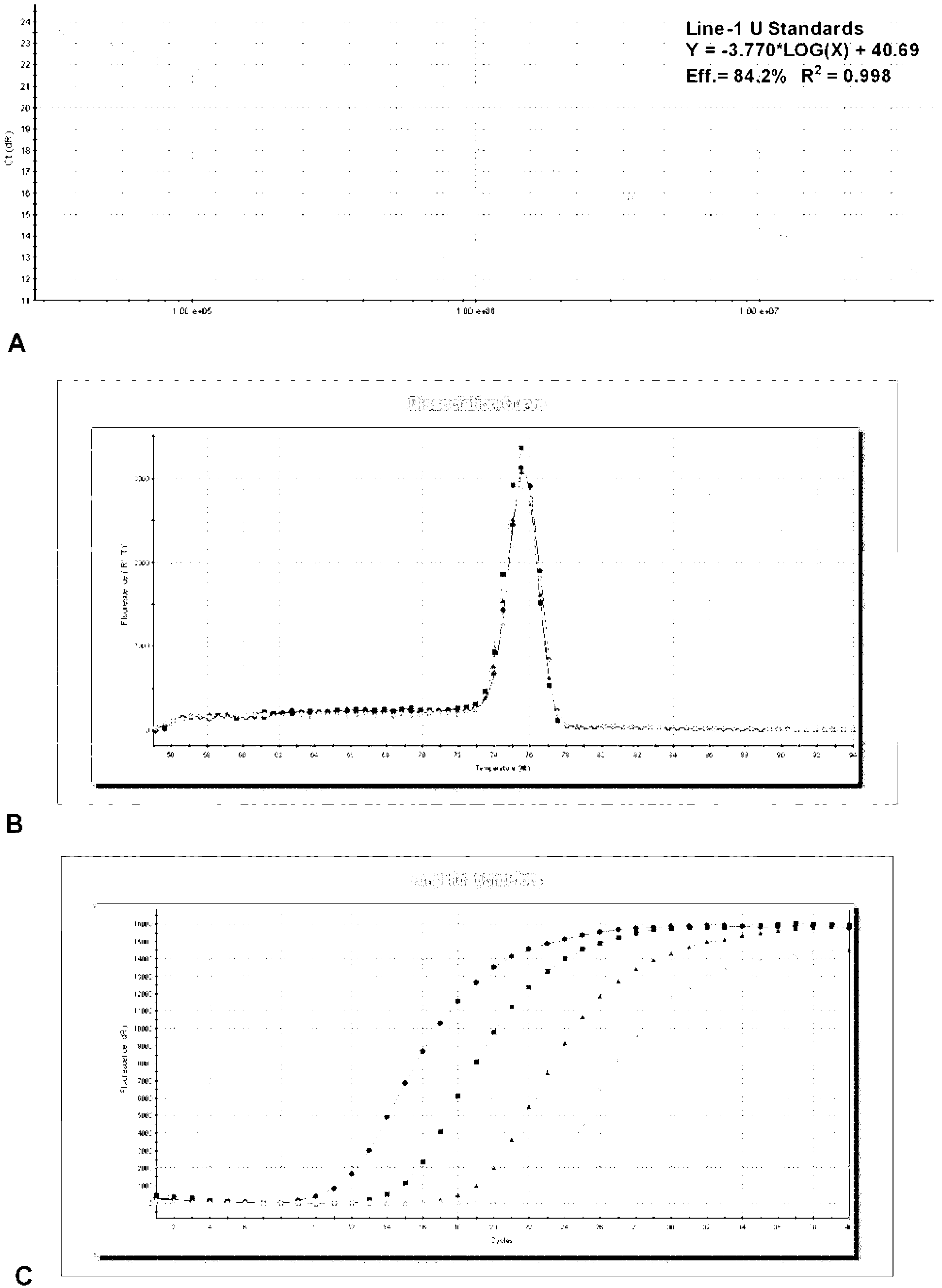
Line-1 gene methylation quantitative detection method
InactiveCN103255210AStrong specificityHigh sensitivityMicrobiological testing/measurementFluorescenceMethylation
The invention discloses a Line-1 gene methylation quantitative detection method, which comprises the steps of: (1) treating a to-be-detected sample to obtain a sample template; (2) designing and synthesizing Line-1 gene methylation primers and Line-1 gene unmethylated primers; (3) respectively using the methylation primers and the unmethylated primers in step (2) to perform a PCR amplification reaction on the sample template in step (1) so as to obtain corresponding PCR amplification products respectively; (4) treating the PCR amplification products in step (3) respectively to obtain corresponding standard templates; and (5) carrying out real-time fluorescence quantitative PCR detection on the sample template in step (1) and the standard templates in step (4) respectively, and calculating the degree of methylation. The method provided in the invention designs the methylation primers and unmethylated primers specific to the Line-1 gene so as to perform qualitative and quantitative detection on Line-1 gene methylation by means of a MSP technique and a real-time fluorescence quantitative PCR technique.
Owner:常州市肿瘤医院
Microbial strain improvement by a htp genomic engineering platform
ActiveCN111223527AIncrease productionImprove productivitySequence analysisInstrumentsMicroorganismBiochemical engineering
The invention relates to a microbial strain improvement by a HTP genomic engineering platform. The present disclosure provides a HTP microbial genomic engineering platform that is computationally driven and integrates molecular biology, automation, and advanced machine learning protocols. This integrative platform utilizes a suite of HTP molecular tool sets to create HTP genetic design libraries,which are derived from, inter alia, scientific insight and iterative pattern recognition. The HTP genomic engineering platform described herein is microbial strain host agnostic and therefore can be implemented across taxa. Furthermore, the disclosed platform can be implemented to modulate or improve any microbial host parameter of interest.
Owner:ZYMERGEN INC
Microbial strain improvement by a htp genomic engineering platform
ActiveCN108027849AIncrease productionImprove productivitySequence analysisSpecial data processing applicationsMicroorganismBiochemical engineering
The present disclosure provides a HTP microbial genomic engineering platform that is computationally driven and integrates molecular biology automation and advanced machine learning protocols. This integrative platform utilizes a suite of HTP molecular tool sets to create HTP genetic design libraries which are derived from inter alia scientific insight and iterative pattern recognition. The HTP genomic engineering platform described herein is microbial strain host agnostic and therefore can be implemented across taxa. Furthermore the disclosed platform can be implemented to modulate or improveany microbial host parameter of interest.
Owner:ZYMERGEN INC
A kind of siRNA recombination interference vector based on the conserved sequence of J subgroup avian leukosis virus env gene and its preparation method and application
InactiveCN103805635BEffective interferenceReduce economic lossGenetic material ingredientsAntiviralsConserved sequenceAvian leukosis viruses
The invention relates to the technical field of genetic engineering, and provides a J avian leukosis virus subgroup env gene conserved sequence-based siRNA (Small Interfering RNA (Ribonucleic Acid)) recombinant interference carrier. The preparation method comprises the following steps: on the basis that the env gene has important meaning for ALV (avian leukosis virus), designing and synthesizing siRNA by the env gene, building to form a hairpin structure of siRNA, further obtaining annealing double-stranded DNA (Deoxyribonucleic Acid), and connecting the double-stranded DNA with the carrier to build the recombinant interference carrier. After the interference carrier and the virus co-transfect cells are adopted, the interference carrier provided by the invention can effectively interfere the transcription and the duplication of the J avian leukosis virus subgroup in the in-vitro cell and the live chicken, the scientific base and the technical support can be provided for preventing and curing the J avian leukosis virus subgroup, and the economic loss caused by the ALV-J infection in the poultry industry can be reduced.
Owner:SHANDONG AGRICULTURAL UNIVERSITY
A gene related to eggplant verticillium wilt resistance, an acquiring method thereof and applications of the gene
PendingCN106167803AGene enrichment selectionMicrobiological testing/measurementOxidoreductasesSolanum torvumGenetic engineering
A gene related to eggplant verticillium wilt resistance, an acquiring method thereof and applications of the gene are provided. The gene has a nucleotide sequence shown as SEQ ID NO.1 in a sequence table. The method includes 1) extracting total RNA and genome DNA of a solanum torvum sample, 2) designing specific primers (a 3'-RACE primer GSP2 and a 5'-RACE primer GSP1) through adopting cDNA obtained by subjecting the RNA extracted in the step 1) to inverse transcription as templates, and performing RACE-PCR amplification, 3) cloning by adopting the genome DNA extracted in the step 1) as templates and by utilizing specific primers gP1F and gP1R designed based on the gene obtained in the step 2), and 4) subjecting a PCR product obtained in the step 3) to recovering, sequencing and sequence analyzing. The gene and the method are suitable for eggplant verticillium wilt research, plant molecular marker assisted breeding and inheritable character improvement in the genetic engineering field, and lay a primary base for verticillium wilt-resisting molecular breeding.
Owner:YANGZHOU UNIV
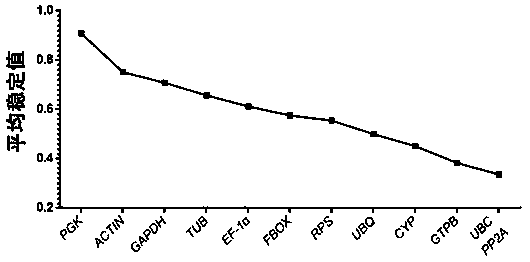
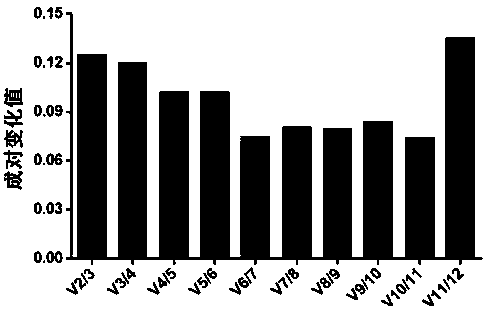
Internal reference gene of cordyceps militaris mycelium under cold stress, and primers, screening method and application of internal reference gene
ActiveCN110791585AImprove stabilityHigh amplification efficiencyMicrobiological testing/measurementDNA/RNA fragmentationReference genesTotal rna
The invention discloses an internal reference gene of the cordyceps militaris mycelium under cold stress, and primers, screening method and application of the internal reference gene. The internal reference gene is UBC, a nucleotide sequence is SEQ ID NO: 1, nucleotide sequences of the forward and reverse primers of the UBC are SEQ ID NO: 13-14. The screening method is as follows: (1) selecting 12internal reference genes as candidate internal reference genes, and designing primers; (2) treating cordyceps militaris under cold stress, extracting the total RNA from the mycelium, measuring the concentration of RNA, synthesizing cDNA through reverse transcription of RNA, using cDNA as a template, and analyzing the candidate internal reference genes by using real-time fluorescence quantitativePCR; and (3) performing Ct value analysis on the real-time fluorescence quantitative PCR data to screen the stable internal reference gene. The application is that the UBC is used as the internal reference gene, and the relative expression of the target gene under cold stress is detected by using real-time fluorescence quantitative PCR. The internal reference gene has good stability. The method issimple, fast, and low in cost.
Owner:CENTRAL SOUTH UNIVERSITY OF FORESTRY AND TECHNOLOGY
DNA library for detecting and diagnosing pathogenic genes of multiple cafe-au-lait macules related diseases and application thereof
PendingCN110724739AAccurate diagnosisKeep healthyMicrobiological testing/measurementProtein nucleotide librariesAtaxia-telangiectasiaA-DNA
The invention relates to a DNA library for detecting and diagnosing pathogenic genes of multiple cafe-au-lait macules related diseases and the application thereof. The library includes 51 pathogenic genes of multiple cafe-au-lait macules related diseases. According to the invention, 51 pathogenic genes of multiple cafe-au-lait macules related diseases are selected, a probe pool is designed, and atarget region library for the 51 pathogenic genes of multiple cafe-au-lait macules is established. The library is used to perform sequencing and find pathogenic mutations by a high-throughput sequencing technology, provides genetic and molecular biological basis for clinical diagnosis, and is accurate, fast, flexible and low-cost. The 51 gene detection regions involved in the invention can detecta variety of multiple cafe-au-lait macules related diseases such as genetic pigment abnormality, Cowden syndrome, ataxia telangiectasia, Noonan syndrome, etc. The DNA library provided by the inventionhas important significance and clinical value for the etiology analysis and differential diagnosis of multiple cafe-au-lait macules.
Owner:福州福瑞医学检验实验室有限公司
A high-throughput (HTP) genomic engineering platform for improving saccharopolyspora spinosa
PendingCN110914425AShorten the timeBacteriaVector-based foreign material introductionEngineeringGenus Saccharopolyspora
The present disclosure provides a HTP microbial genomic engineering platform for Saccharopolyspora spp. that is computationally driven and integrates molecular biology, automation, and advanced machine learning protocols. This integrative platform utilizes a suite of HTP molecular tool sets to create HTP genetic design libraries, which are derived from, inter alia, scientific insight and iterativepattern recognition.
Owner:ZYMERGEN INC
A chip for microbial oil and gas exploration and its application
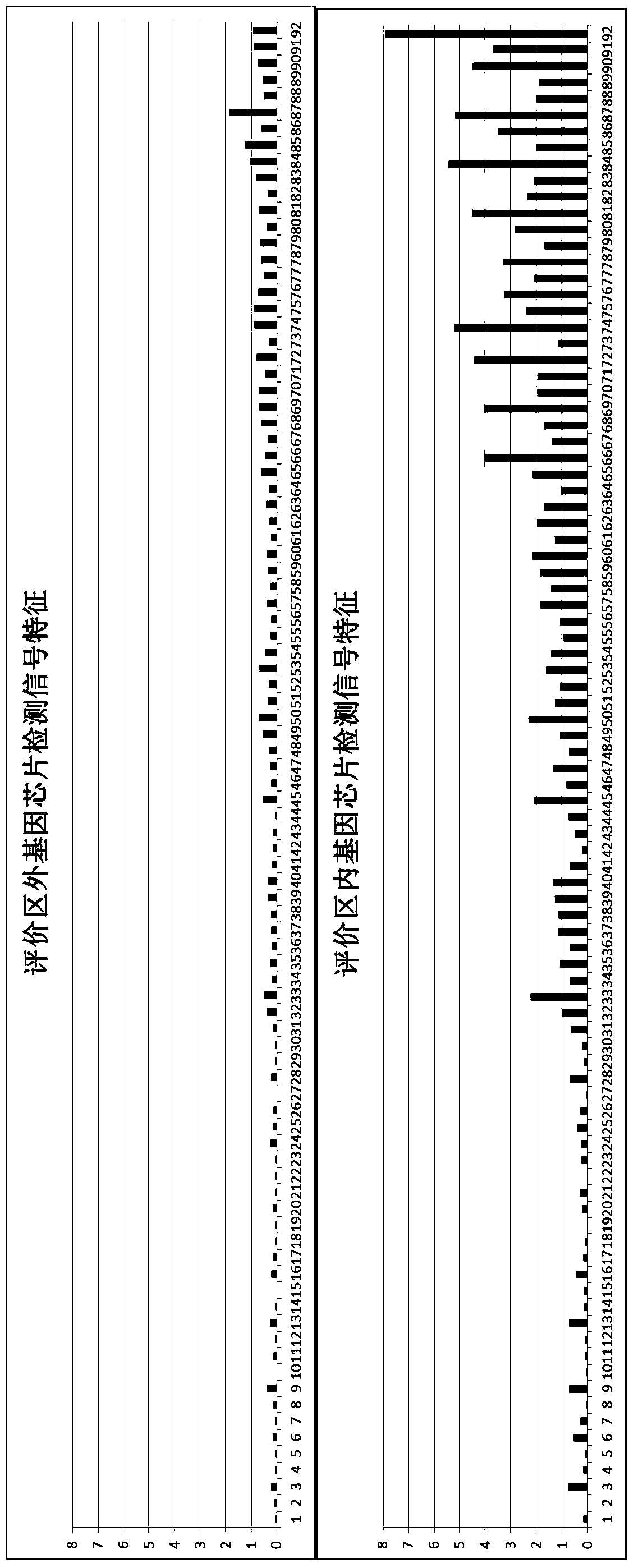
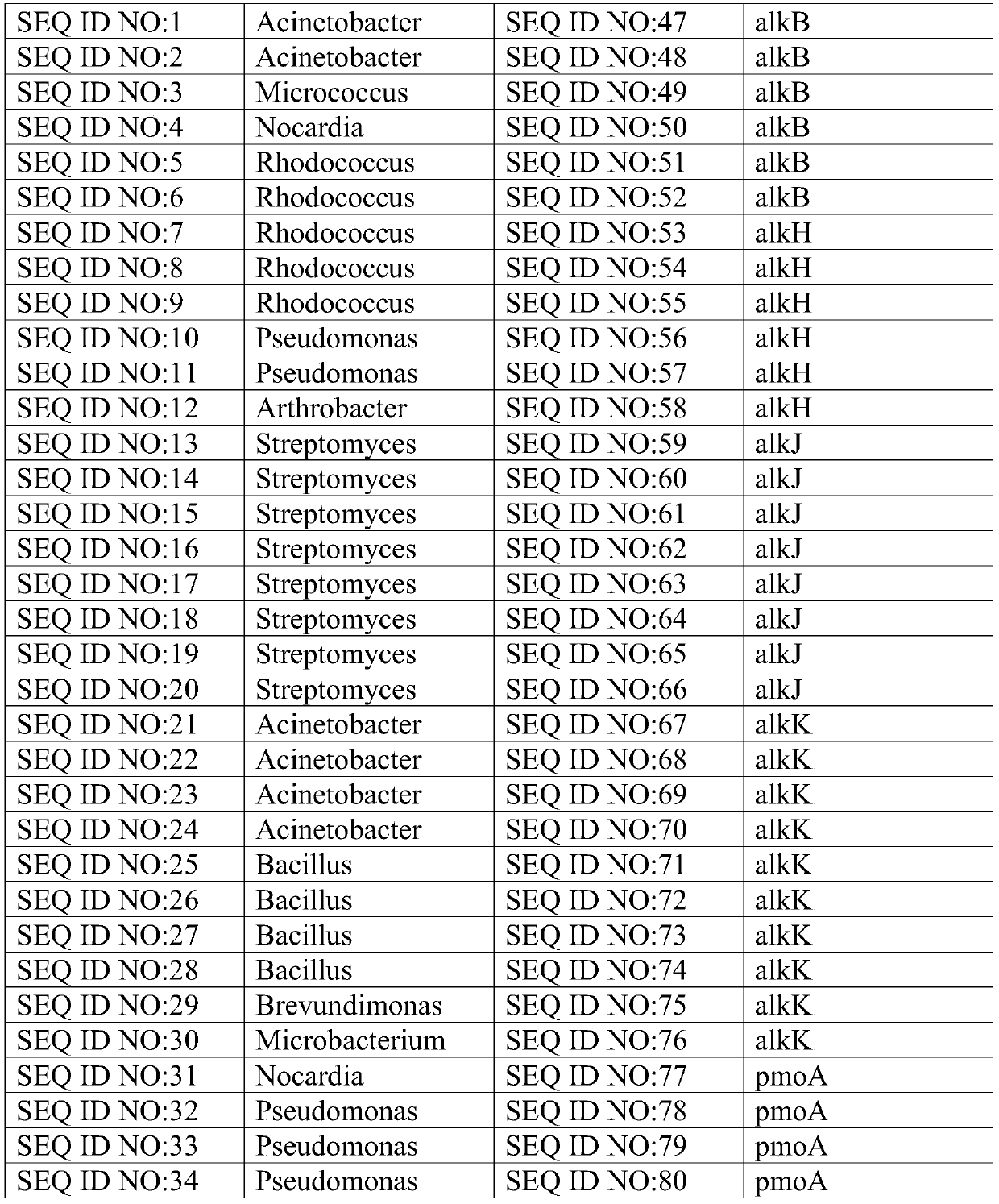
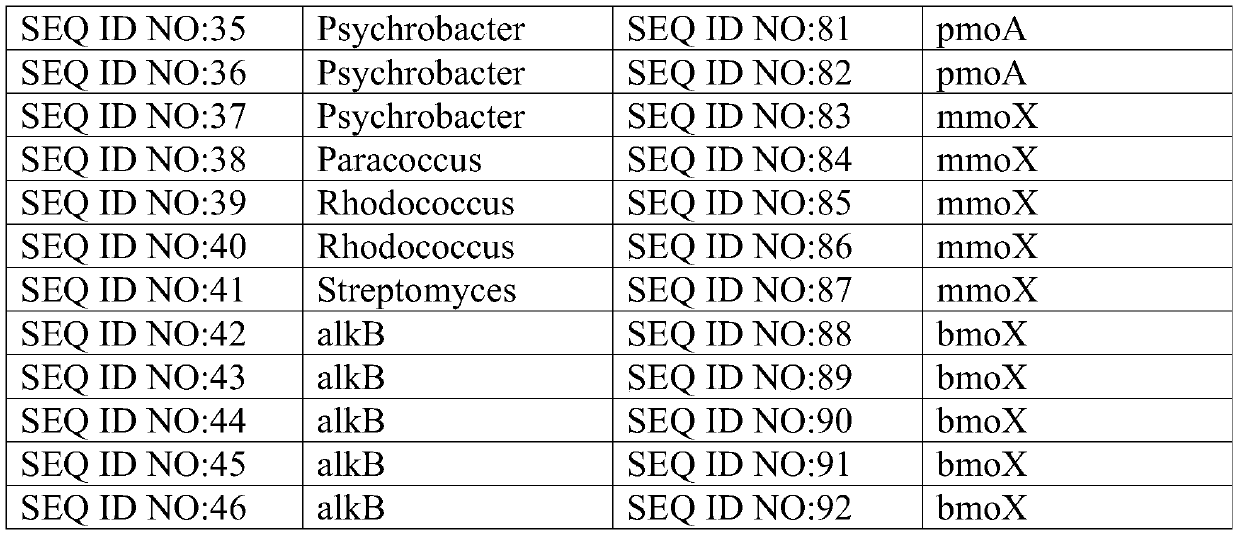
ActiveCN106011241BReduce incubation timeStrong specificityMicrobiological testing/measurementMicroorganism based processesMicrobial oilFlora
The invention relates to the technical field of biological detection and particularly relates to a chip for microbial prospecting of oil and gas and application thereof. The invention provides a probe set, a chip and a kit for prospecting of oil and gas and a prospecting method. The probe set has good specificity according to the design of a microbial 16s rRNA gene as well as an alkB gene, an alkH gene, an alkJ gene, an alkK gene, a pmoA gene, an mmoX gene or a BmoX gene. By applying the primer set or the chip to prospecting of oil and gas resources, the flora culture time of the conventional method is saved, the operation is simple and convenient, and the accuracy is high. A more reliable basis can be provided for prospecting of oil and gas resources.
Owner:ADVANCED ENERGY & ENVIRONMENTAL TECH INC
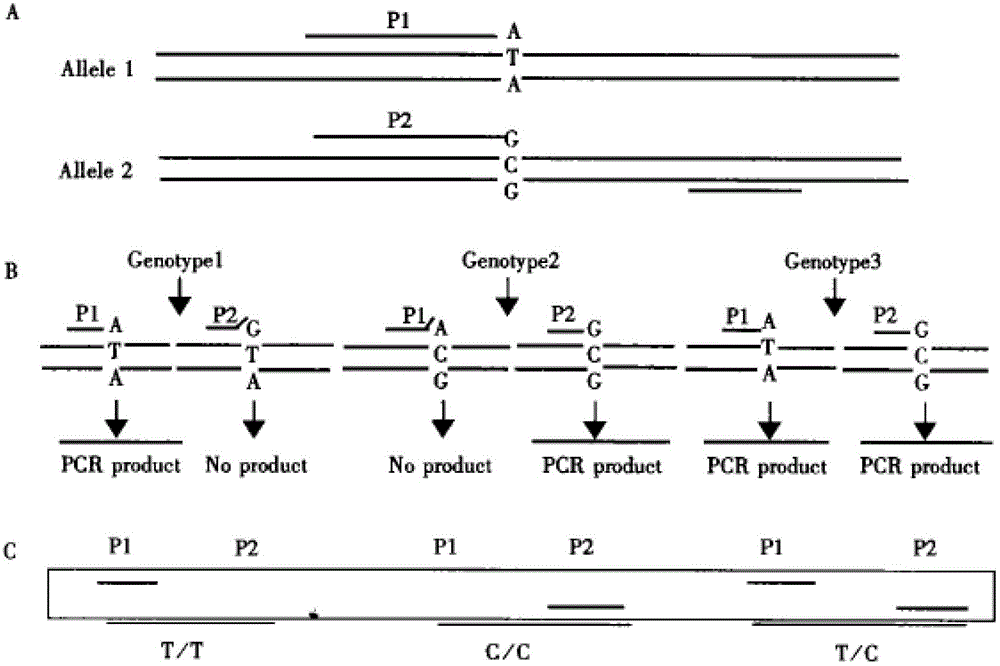
Method for designing SNP (single-nucleotide polymorphism) molecular marker with base substitution or insertion deletion
InactiveCN103146823AEasy to knowEfficient use ofMicrobiological testing/measurementInsertion deletionAgricultural science
The invention discloses a method for designing an SNP (single-nucleotide polymorphism) molecular marker with base substitution or insertion deletion. Since common TaqDNA polymerase needs normal amplification, 5' does not need to be completely paired with the template, and 3' needs complete pairing. Thus, the discovered SNP site can be specifically designed in the 3' position of the primer; the primer designed according to the mutant gene and the wild gene can form 3' terminal mispairing, resulting in incapability of normal amplification; and similarly, the primer designed according to the wild gene and the 3' terminal of the mutant gene generate mispairing, resulting in incapability of normal amplification in the mutant individual genome. By carrying out PCR (polymerase chain reaction) twice and detecting the PCR result by a specific technique, the invention can conveniently determine the genotype of the individual SNP site.
Owner:NORTHWEST A & F UNIV
Polyepitope hepatitis C antigen complex polypeptide vaccine and its prepn and application
InactiveCN1347735AEffectively induces a humoral immune responseImproving immunogenicityAntibody medical ingredientsFermentationAntigenHepatitis C vaccine
The present invention relates to molecular biological technology and aims at providing one kind of immunogenic hepatitis C vaccine to HCV virus with serious harm. The said polyepitope antigen complexpolypeptide consists of 131 amino acids and has molecular weight of 13.8 KD and isopotential point of 8.3. Its preparation includes the steps of gene design, effective expression, separation and purification of expression product and preparation of polypeptide immunogen. Test data shows that the vaccine of the present invention has excellent scene of being used as hepatitis C preventing vaccine and detection and diagnosis material.
Owner:楚雄老拨云堂药业有限公司
Vibrio shilonii multiple virulence factor GeXP rapid detection kit and detection method thereof
ActiveCN102899402AImprove accuracyWide detection coverageMicrobiological testing/measurementAgainst vector-borne diseasesMicrobiologyMultiplex pcrs
The present invention discloses a Vibrio shilonii multiple virulence factor GeXP rapid detection kit and a detection method thereof. According to the present invention, genome DNA of Vibrio shilonii is extracted and is adopted as a template, 13 pairs of multi-gene PCR primers and a pair of universal primers are adopted to carry out multiplex PCR amplification to obtain multiplex PCR reaction products, and a GenomeLab GeXP genetic analysis system is adopted to carry out detection analysis. With the present invention, the special primers are designed based on the 13 Vibrio shilonii virulence genes, a single-tube reaction is performed to rapidly and efficiently detect the 13 genes in one time so as to obtain the following results, the detection coverage is wide, and the Vibrio shilonii detection accuracy can be substantially improved, wherein the results comprise whether the detected sample contains the virulence carrying gene and the Vibrio shilonii having the potentially pathogenic ability. In addition, a certain theory basis is provided for coral bleaching phenomenon prevention and control, and important practical significances are provided for further coral island protection in our country and further degradation prevention.
Owner:SOUTH CHINA SEA INST OF OCEANOLOGY - CHINESE ACAD OF SCI
Gene chip detection method for pathogenic bacteria in sea water of bathing place
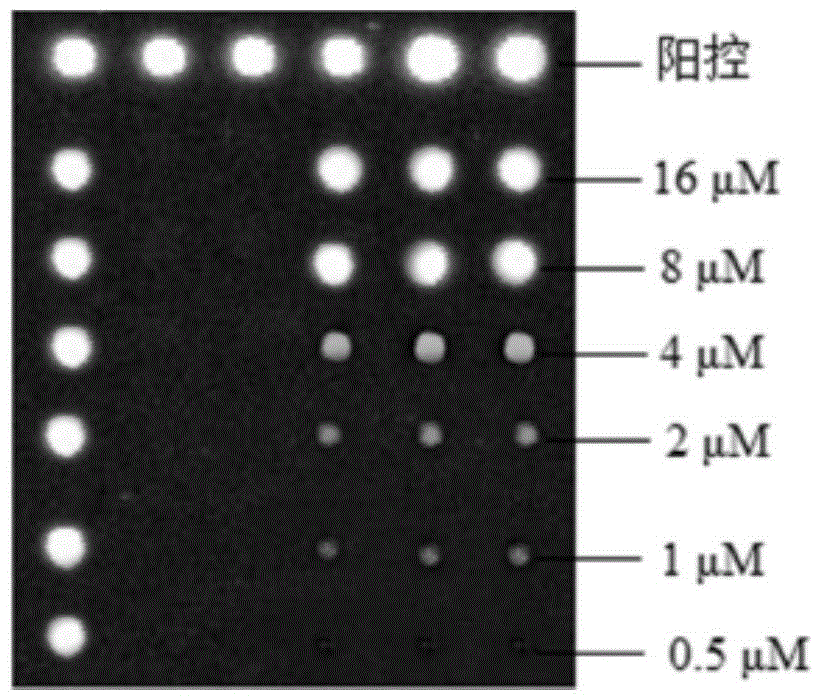
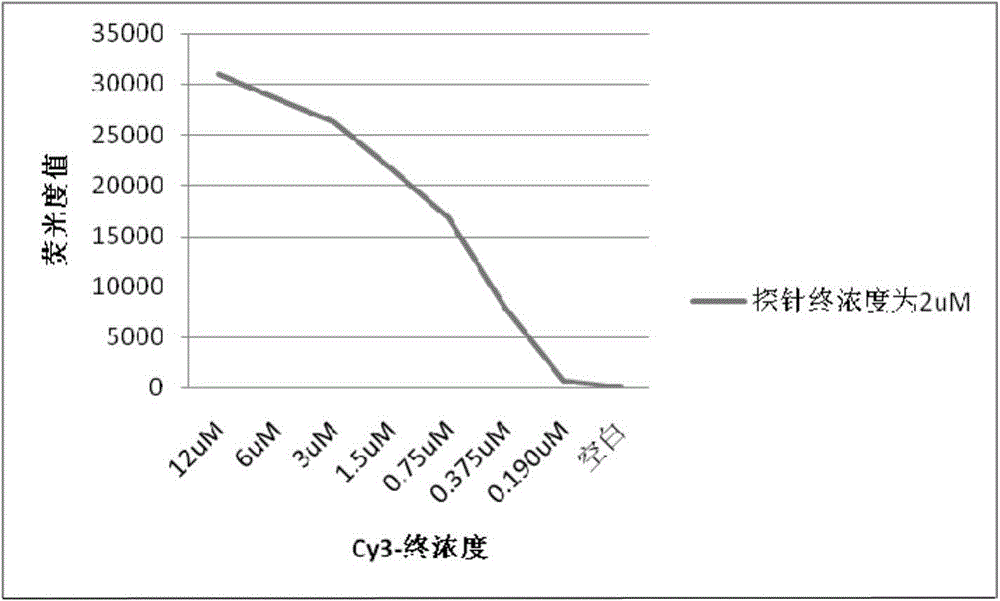
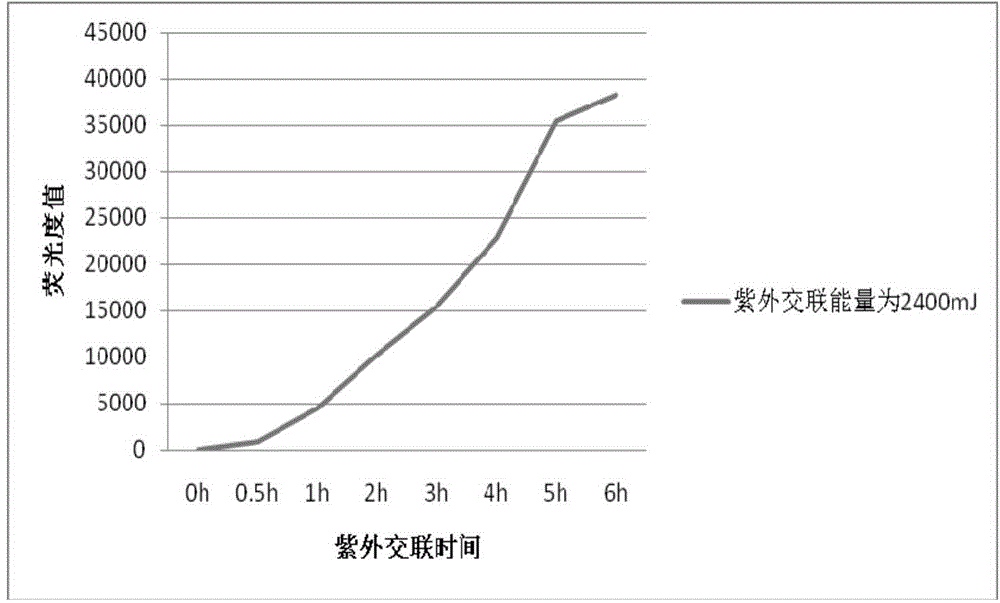
InactiveCN104962617AProtect healthPromote sustainable developmentMicrobiological testing/measurementAgainst vector-borne diseasesBathing BeachesFluorescence
The invention belongs to the technical field of environment detection and relates to a gene chip detection method for pathogenic bacteria in sea water of a bathing place and the detection method is used for simultaneously detecting main pathogenic bacteria in sea water of the bathing place. According to the invention, specific primers and specific probes are designed according to specific target genes, PCR products are labeled through fluorescence, and the purpose of quickly detecting pathogenic bacteria in a high-flux manner is realized through the steps of preparing an amino chip, hybridizing, cleaning, scanning and the like; and in addition, probe concentration, Cy3 random primer concentration, and chip ultraviolet crosslinking time are optimized. By adopting the method, the environmental status and variation tendency of a bathing beach can be comprehensively mastered with a short time and sensitive, accurate and efficient detection of pathogenic bacteria in sea water of the bathing place can be realized.
Owner:NATIONAL MARINE ENVIRONMENTAL MONITORING CENTRE
CHO cell strain modified based on CRISPR/Cas9 gene editing and preparation method thereof
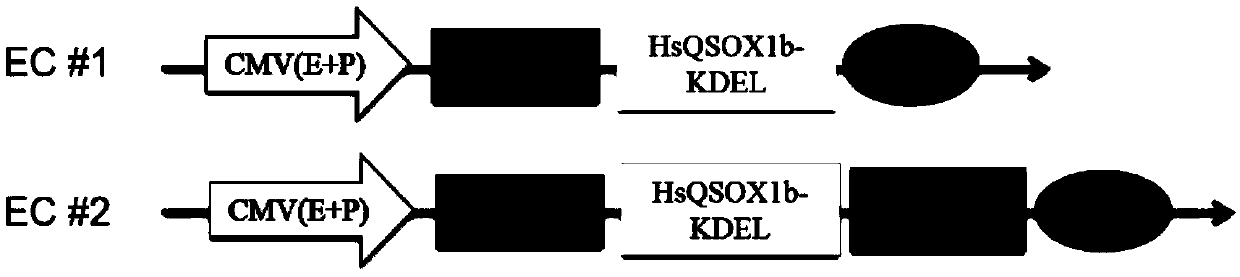
PendingCN110408596AEasy to transformRetrofit smallGenetically modified cellsOxidoreductasesSurvivinProtein C
The invention provides a CHO cell strain modified based on CRISPR / Cas9 gene editing and a preparation method thereof. Sulfhydryl oxidase (HsQSOX1b) related to disulfide bond folding and survivin related to apoptosis are selected as target genes and designed as heterogenous expression cassettes EC#1 and EC#2, and a novel CRISPR / Cas9 gene editing technology is used for integrating the heterogenous expression cassettes into a specific genetic locus of a CHO cell. According to the CHO cell strain modified based on CRISPR / Cas9 gene editing and the preparation method thereof, the CRISPR / Cas9 gene editing technology is used, human HsQSOX1b and survivin genes accurately edit a CHO-K1 cell, a rapid and efficient host cell modification technology platform is established, a monoclonal cell strain with high anti-apoptosis ability and high disulfide bond catalytic efficiency and protein expression quality is obtained, and a theoretical and technical support is provided for modification of an efficient antibody cell strain which meet the requirements of industrial production.
Owner:EAST CHINA UNIV OF SCI & TECH
Alfalfa CRISPR/Cas9 genome editing system and application thereof
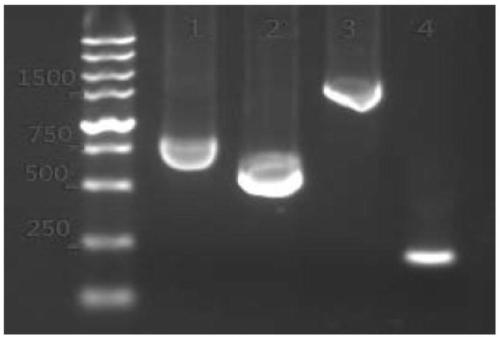
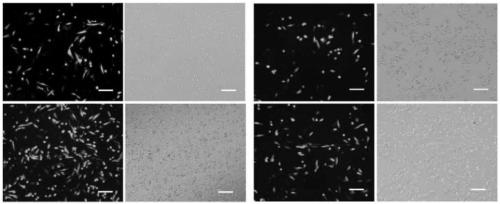
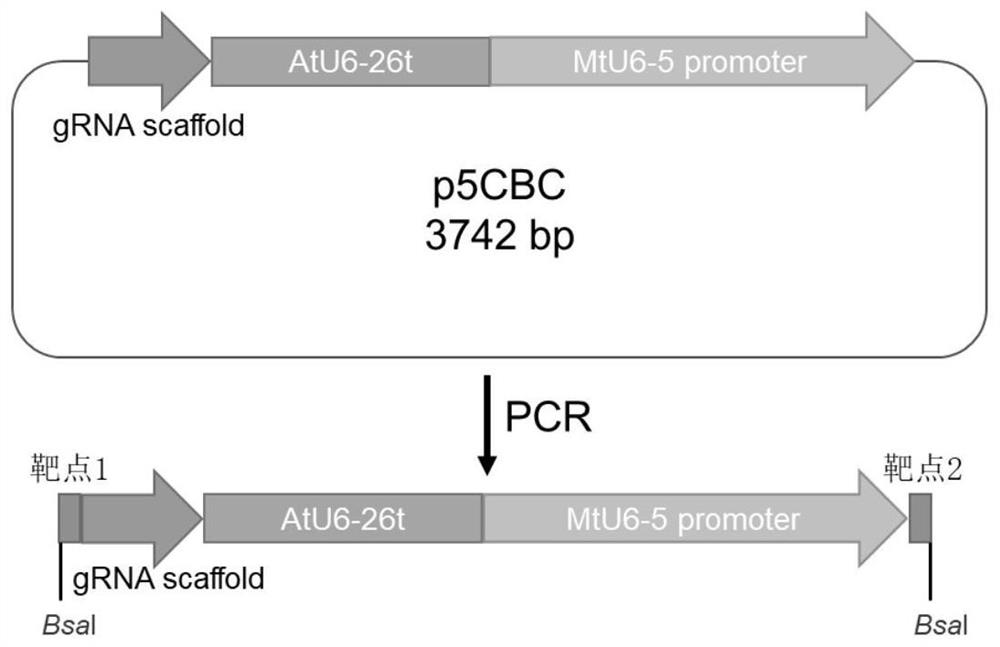
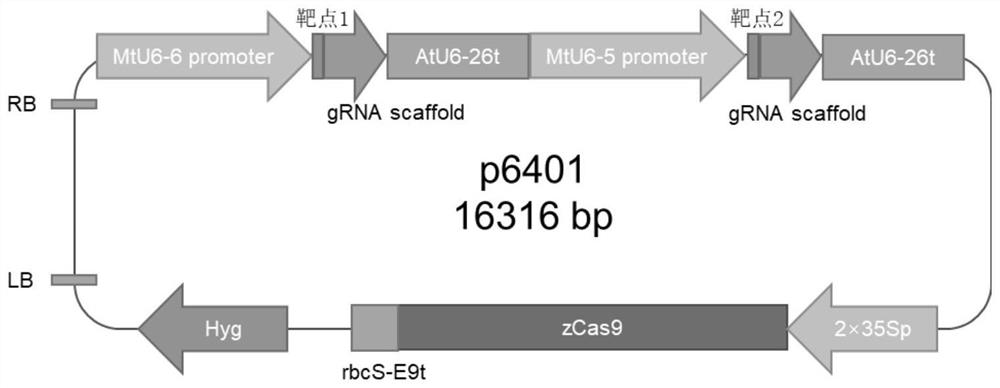
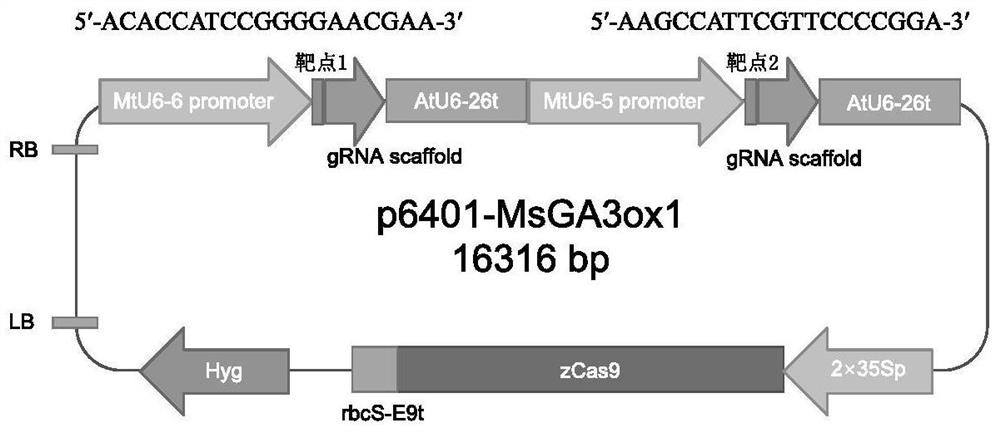
ActiveCN113493803AEfficiently obtainedImprove editing efficiencyHydrolasesPlant peptidesGenome editingGene drive
The invention discloses application of a CRISPR / Cas9 genome editing system in alfalfa gene editing, the CRISPR / Cas9 genome editing system comprises an expression vector, the expression vector can comprise a Cas9 expression cassette, an sgRNA1 expression cassette and an sgRNA2 expression cassette, and the Cas9 expression cassette is used for expressing Cas9. The sgRNA1 expression cassette is used for expressing sgRNA with the name of sgRNA1, and the sgRNA2 expression cassette is used for expressing sgRNA with the name of sgRNA2; the sgRNA1 expression cassette contains a promoter with the name of MtU6-6promoter and a sgRNA1 gene driven by the MtU6-6promoter, and the sgRNA2 expression cassette contains a promoter with the name of MtU6-5promoter and a sgRNA2 gene driven by the MtU6-5promoter; the sgRNA1 and the sgRNA2 can aim at the same target gene or different target genes of the alfalfa. According to the invention, double targets are designed for a target gene according to the characteristics of an autotetraploid of alfalfa, a gene editing binary vector p6401-Target is constructed, and alfalfa is subjected to agrobacterium-mediated transformation to obtain a regenerated plant. According to the optimized genome editing system, the alfalfa gene editing efficiency is greatly improved, the plant gene editing efficiency can reach up to 100%, and the single target editing efficiency can reach up to 96.9%.
Owner:CHINA AGRI UNIV
Primers for detecting exogenous genes in transgenic rice TT51-1 derivative line and method thereof
PendingCN110791581ASimple or notSimple homozygosityMicrobiological testing/measurementDNA/RNA fragmentationForward primerGenetically modified rice
The invention discloses primers for detecting exogenous genes in a transgenic rice TT51-1 derivative line and a method thereof. A forward primer P1, a forward primer P3 and a reverse primer P2 are designed for an exogenous gene cry1Ab / cry1Ac in the transgenic rice TT51-1 derivative line, during detection, the three primers are adopted to perform multiplex PCR amplification on genomic DNA of TT51-1and progeny materials derived from TT51-1 in a same PCR system, and then the banding pattern of an amplification product is detected. According to the design, simple and efficient detection of existence and homozygosity of exogenous genes is realized.
Owner:INST OF FOOD CROPS HUBEI ACAD OF AGRI SCI
Improvement of microbial strains using the htp genome engineering platform
ActiveCN108027849BIncrease productionImprove productivitySequence analysisInstrumentsMicrobial GenomesMicroorganism
The present invention provides a computer-driven HTP microbial genome engineering platform that integrates molecular biology, automation and advanced machine learning solutions. The integrated platform uses a set of HTP molecular tools to create HTP genetic design libraries, which are obtained using inter alia scientific insights and iterative pattern recognition. The HTP genome engineering platform described herein is microbial strain host agnostic and thus enables construction across taxa. Additionally, the disclosed platform can be constructed to modulate or improve any microbial host parameter of interest.
Owner:ZYMERGEN INC
A Gene Knockout Method for Rapidly Acquiring Large Deletions in Cell Lines Using the CRISPR/Cas9 System
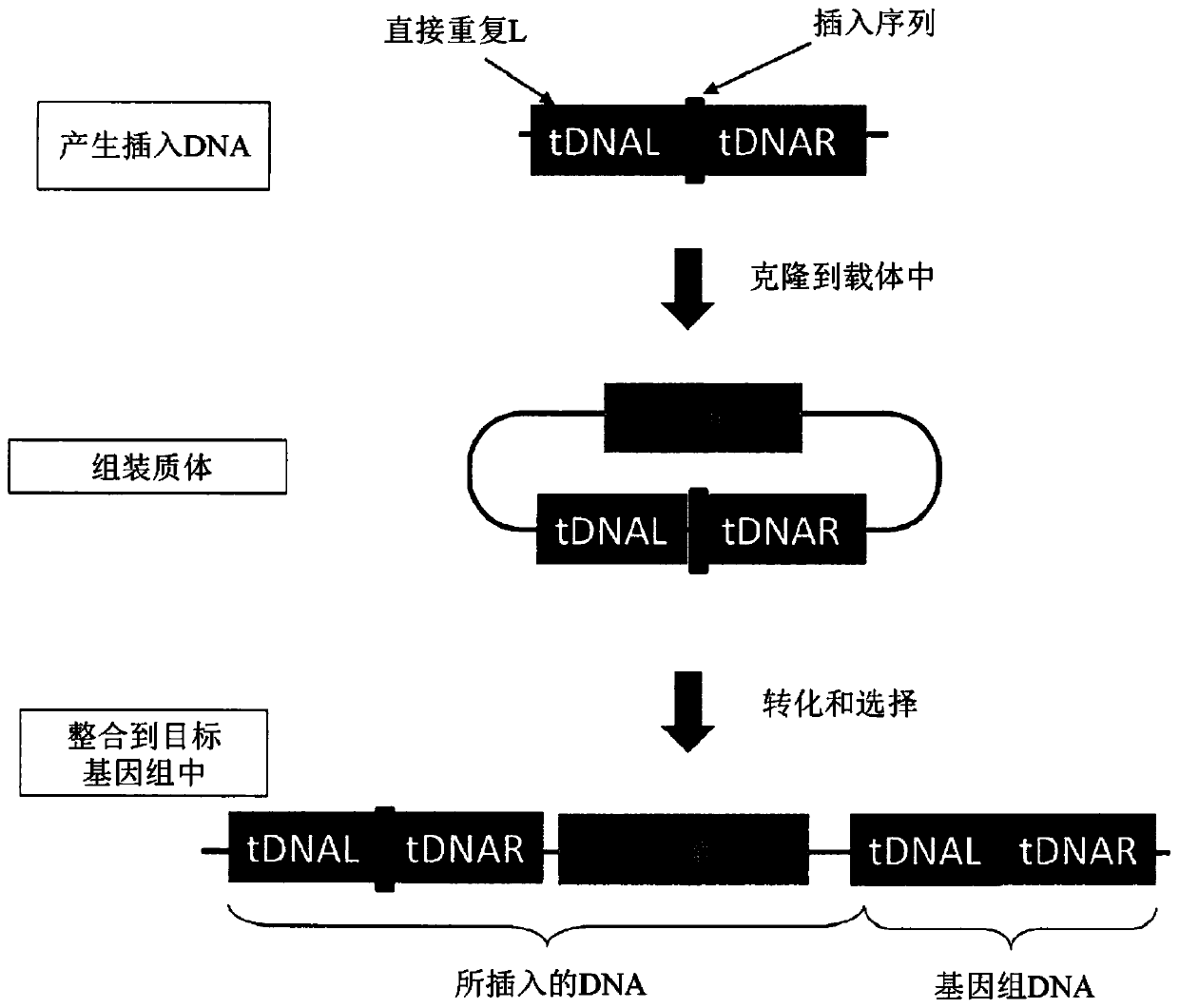
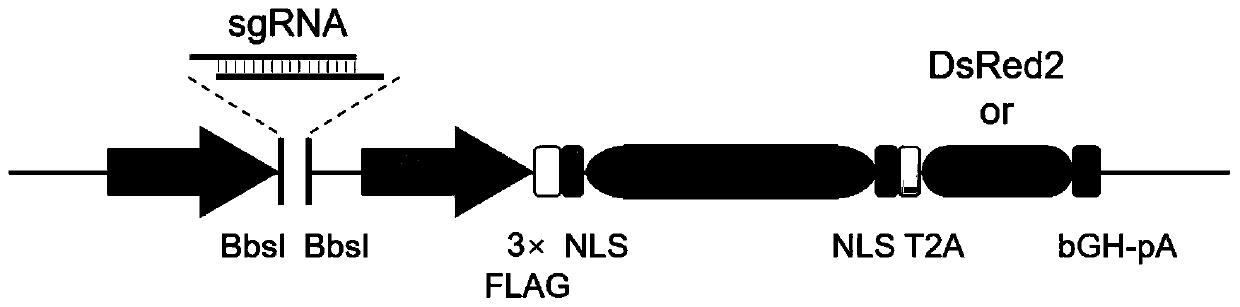
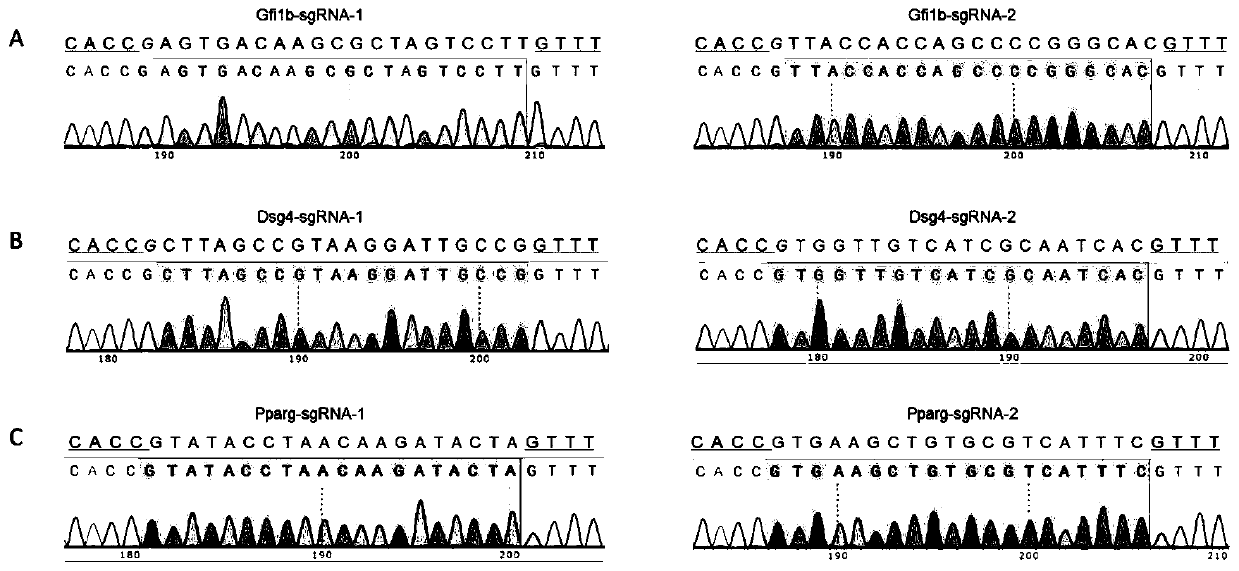
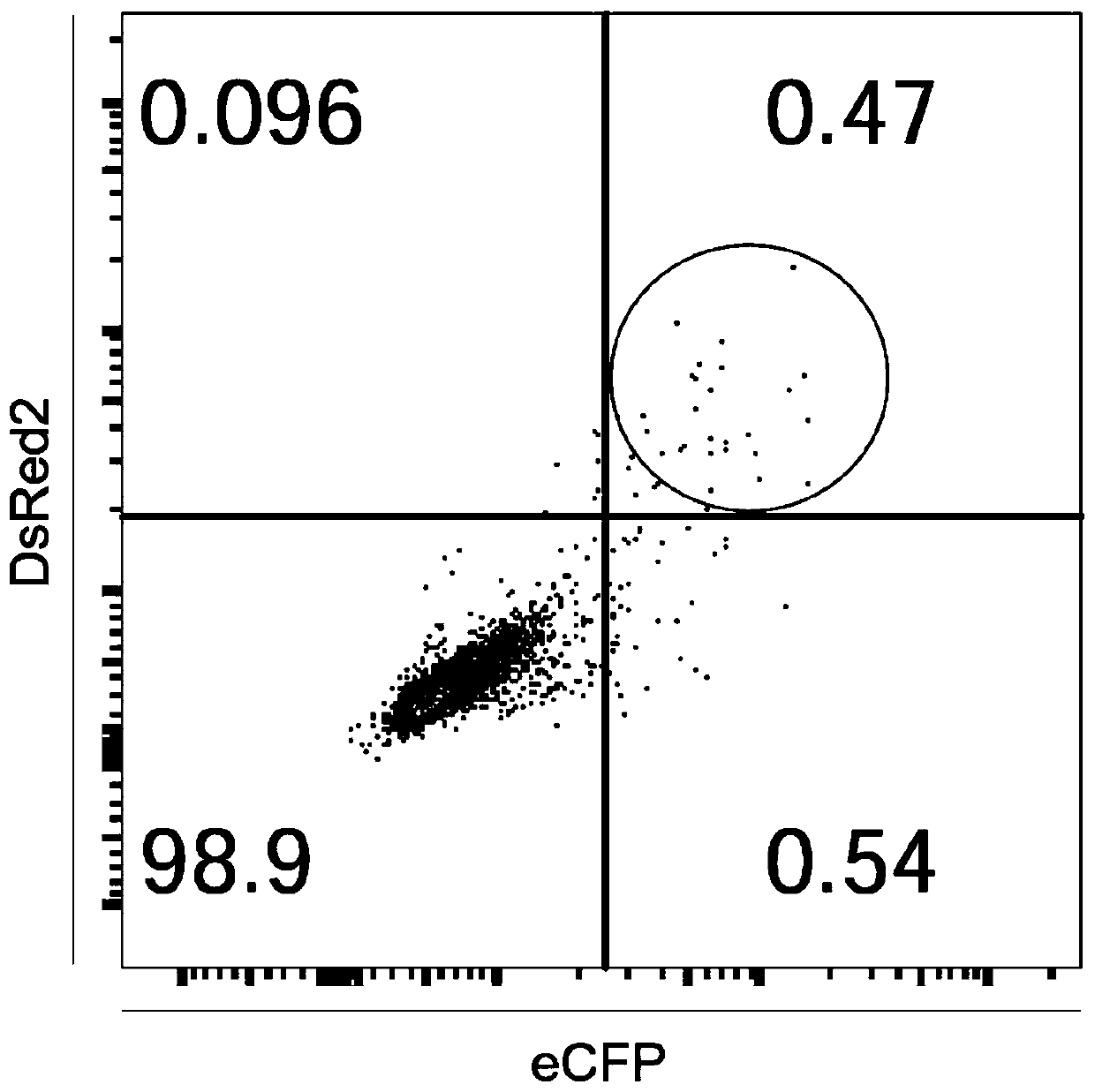
ActiveCN107435051BImprove editing efficiencyExcitation wavelength farVectorsStable introduction of DNAGel electrophoresisGenetic engineering
The invention relates to a cell line gene knockout method for rapidly obtaining large fragment deletion through a CRISPR / Cas9 system, and belongs to the fields of genetic engineering and genetic modification. In the present invention, the pX458 vector is modified to carry DsRed2 and ECFP, and then multiple specific sgRNA sites are designed for the target gene, which are connected to the modified vector. Sorting, single cells with edited genomes can be obtained very quickly, and then single-cell DNA sequences are amplified by PCR, and single cells with large deletions can be selected by gel electrophoresis. By combining the CRISPR / Cas9 system, flow cytometry single cell sorting and fluorescent protein screening on the expression vector, the present invention can obtain a positive single clone with large fragment deletion in a short time, and greatly improve the cell line Gene knockout work efficiency.
Owner:XINXIANG MEDICAL UNIV
Method for rapidly detecting chicken fast and slow feathering phenotypes by multiple PCR system
PendingCN113699245AEasy to operateReduce workloadMicrobiological testing/measurementDNA/RNA fragmentationMolecular identificationMedicine
The invention provides a molecular identification method for chicken fast and slow feathering phenotypes. A primer K-R1: SEQ ID NO.1, a primer K-F1: SEQ ID NO.2 and a primer K-R2: SEQ ID NO.3 which are designed for a PRLR gene and an SPEF2 gene are used for amplification. Compared with a traditional PCR method, the three-primer PCR method has the advantages that the cost is saved, the false negative rate is low, and the judgment is accurate.
Owner:CHINA AGRI UNIV
Genetic design method of spiking neural P system
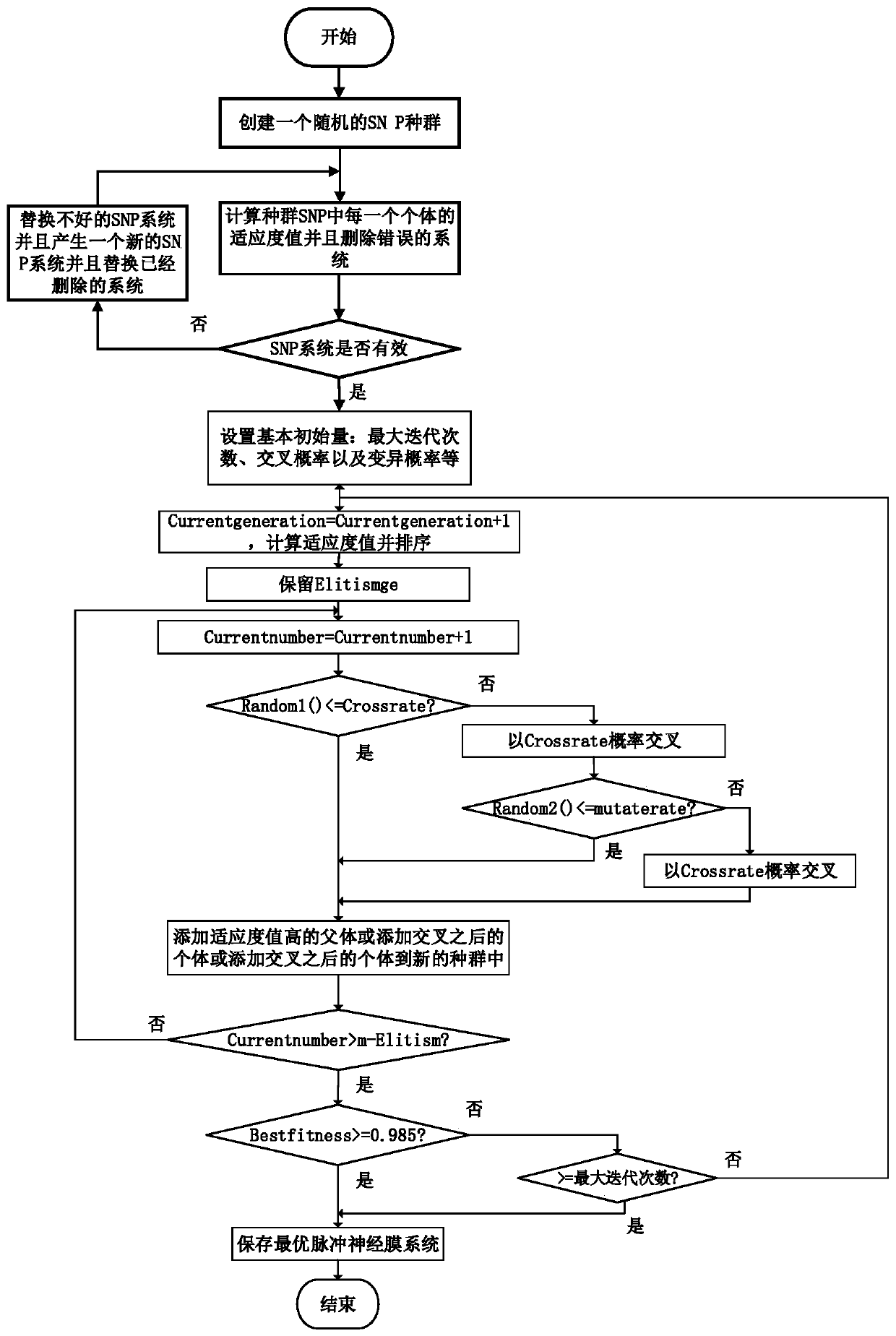
InactiveCN110909866ARealize automatic optimizationAvoid easy mistakesNeural architecturesPhysical realisationGenetics algorithmsP system
The invention discloses a genetic design method of a spiking neural P system. The genetic design method comprises the steps of randomly generating the spiking neural P system to form a population; calculating the fitness value of each spiking neural P system in the population; replacing the spiking neural P system of which the fitness value is smaller than a fitness threshold value or the fitnessvalue is a null value; and designing all spiking neural membrane systems in the population by adopting a genetic algorithm. Compared with a traditional manual design method, the method has the advantages that a unified fitness function is provided to serve as the basis of population evolution, the genetic algorithm is applied to search for the optimal spiking neural P system in the initial population, and automatic optimization of the spiking neural P system is achieved. According to the method, errors easily occurring in the manual derivation process can be effectively avoided, and the time required for designing the spiking neural P system model is greatly shortened.
Owner:SOUTHWEST JIAOTONG UNIV
Rapid typing identification method of lycium barbarum S gene based on targeted sequencing
ActiveCN113403415ARapid typing identificationHigh detection throughputMicrobiological testing/measurementDNA/RNA fragmentationSequence analysisGenotyping
The invention relates to a rapid typing identification method of lycium barbarum S genes based on targeted sequencing. The rapid typing identification method comprises the following steps of designing probes according to S genes of a plurality of reference lycium barbarum varieties, carrying out hybrid capture and sequencing analysis on genes of a lycium barbarum sample to be detected by utilizing a designed probe composition, and comparing a sequencing result with the S genes of the referencelycium barbarum varieties, and judging whether the S gene of the to-be-detected lycium barbarum sample is matched with the S gene of the reference lycium barbarum varieties, and determining the reference lycium barbarum varieties matched with the S gene of the to-be-detected lycium barbarum sample. Specific amplification primers are designed for a plurality of SNP loci to be detected, and the rapid typing identification of the lycium barbarum S-RNase gene is realized by using a genotyping by target sequencing (GBTS) technology.
Owner:WOLFBERRY SCI INST NINGXIA ACAD OF AGRI & FORESTRY SCI
Features
- R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
Why Patsnap Eureka
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Social media
Patsnap Eureka Blog
Learn More Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com