Amplification internal standard preparation based on DNA stochastic shuffling technology
A technique for amplifying internal standards and techniques, applied in DNA preparation, recombinant DNA technology, microorganism-based methods, etc., can solve problems such as no reports of amplification internal standard preparation methods, and overcome the impact of PCR amplification efficiency , to ensure consistency, accurate quantitative effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment
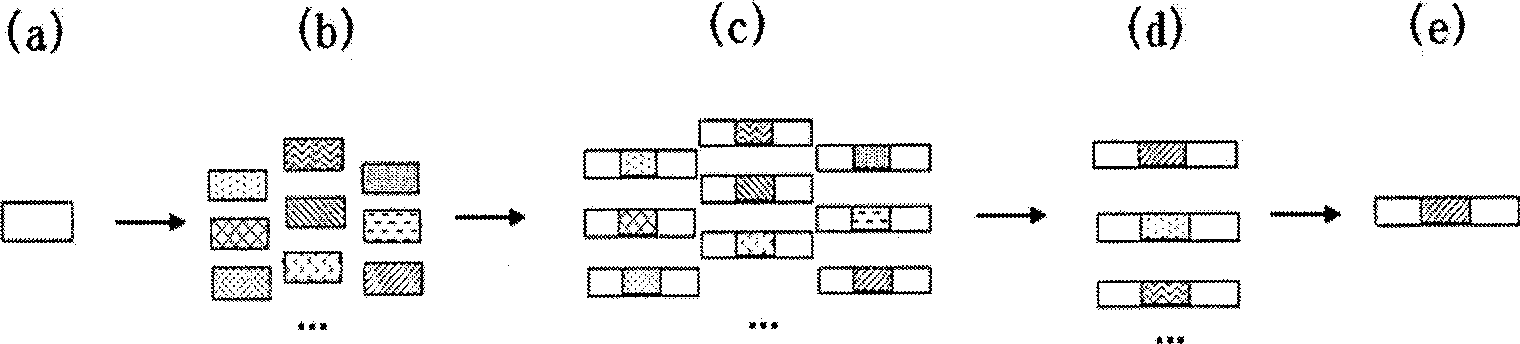
[0024] figure 1 Schematic diagram of the design steps for the amplification internal standard sequence, in the figure: (a) is the design of the target fragment probe; (b) is the random shuffling of the sequence of the target fragment probe binding site; (c) is the internal standard fragment to be screened and amplified (d) is the screening of the amplified internal standard fragment; (e) is the determination of the amplified internal standard sequence.
[0025] Step 1: Analyze the target gene and design specific detection primers and TaqMan probes
[0026] The known specific genes of Listeria monocytogenes were analyzed, and the target gene hlyA was selected for detection. Using the public BLAST software in Genbank, the sequence of the hlyA gene was compared with other microorganisms, and a sequence segment with high specificity was selected. Based on this sequence, a pair of primers and probes were designed using the software Bacon Designer 5.0.
[0027] The primer and prob...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com