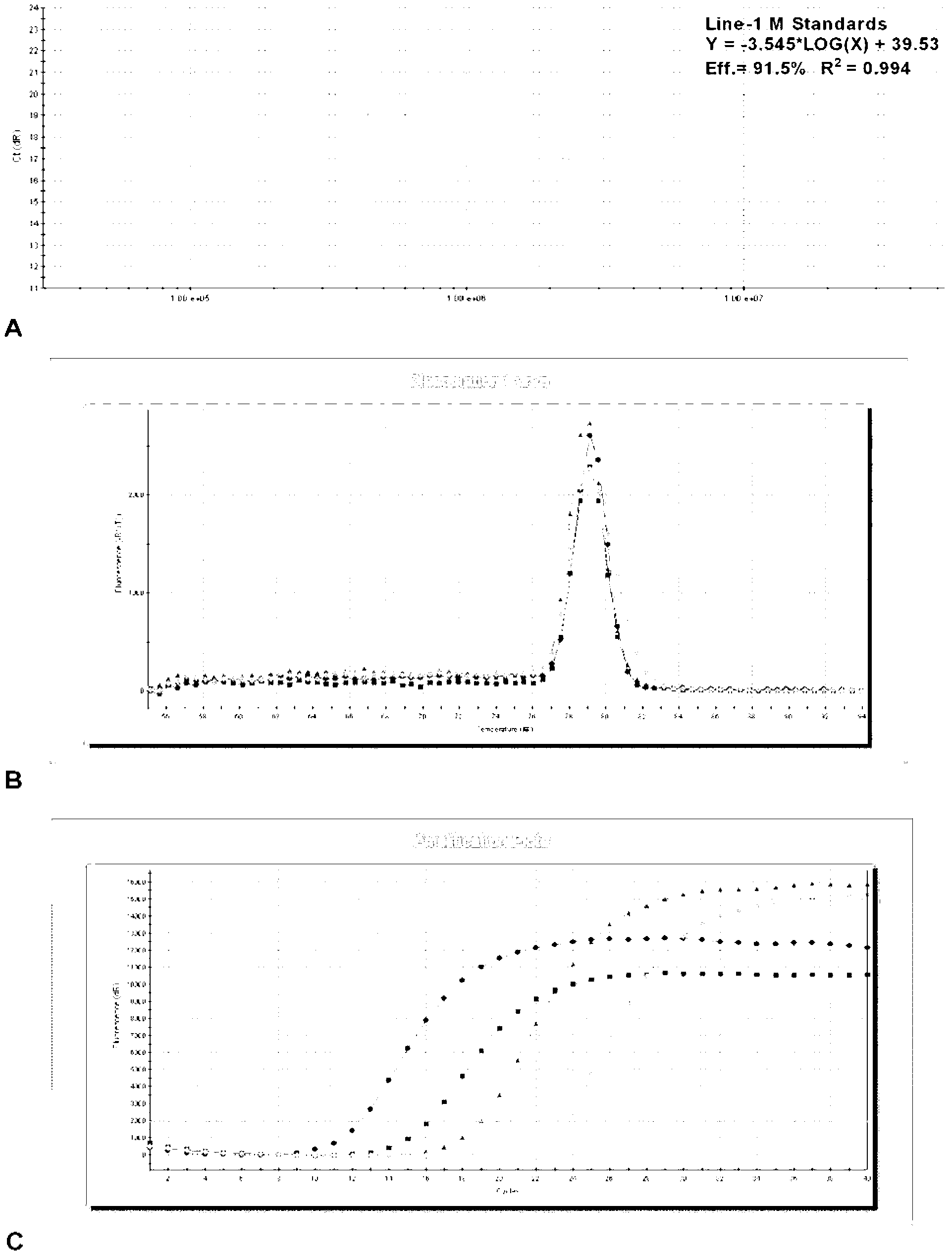
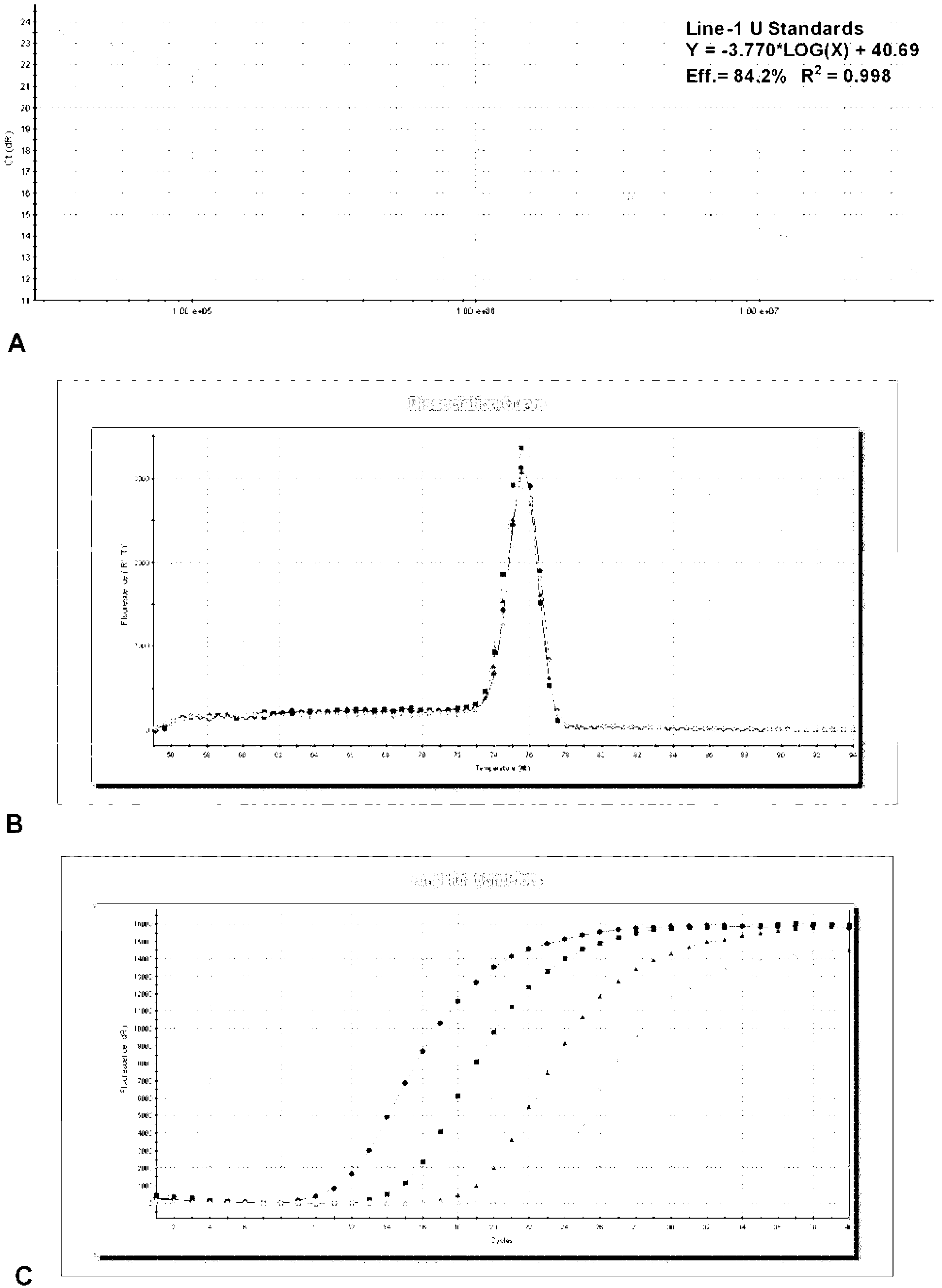
Line-1 gene methylation quantitative detection method
A methylation quantification and line-1 technology, applied in the field of biomedicine, can solve problems such as the method for detecting the methylation of Line-1 gene that has not yet been found, and achieve the effects of good specificity, accurate quantification and high sensitivity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1)
[0037] In this embodiment, the clinical tissue of liver cancer patients is used as the sample to be tested, and the degree of methylation of the Line-1 gene is quantitatively detected.
[0038] The quantitative detection method of the present embodiment has the following steps:
[0039] ① Process the sample to be tested to obtain the sample template.
[0040] A. Extract DNA from the sample to be tested.
[0041] Take 50 mg of fresh liver cancer resected tissue, and extract DNA with a genomic DNA extraction kit (operate according to the instructions) to obtain the DNA of the sample to be tested.
[0042] B. Carry out sodium bisulfite modification to the extracted DNA, the specific process is as follows:
[0043] 1) Dissolve 1 μg of DNA in 20 μL of water and bathe in 95°C water for 10 minutes; immediately put it in an ice bath; add 2 μL of 10M sodium hydroxide to denature the DNA and become single-stranded DNA.
[0044] 2) Add 230 μL of sodium bisulfite modification solution ...
Embodiment 2~ Embodiment 10)
[0094] The clinical tissues of different liver cancer patients were taken as samples to be tested, and the quantitative detection of Line-1 gene methylation was carried out according to the method of Example 1, and the results are still shown in Table 3.
[0095] In the above-mentioned embodiments, liver cancer tissue was used as the sample to quantitatively detect the degree of methylation of the Line-1 gene.
[0096] Blood (whole blood, serum), body fluid, secretion or puncture can also be collected as samples to be tested, and the above method can be used to quantitatively detect the degree of methylation of the Line-1 gene.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com