Accurate method capable of carrying out in-vivo target activity evaluation and off-target detection in batches
A batch, active technology, applied in biochemical equipment and methods, microbial determination/inspection, library creation, etc., can solve the problems of complex methods and low accuracy, and achieve improved detection throughput, library reduction, The effect of accurate detection results
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
specific Embodiment approach 1
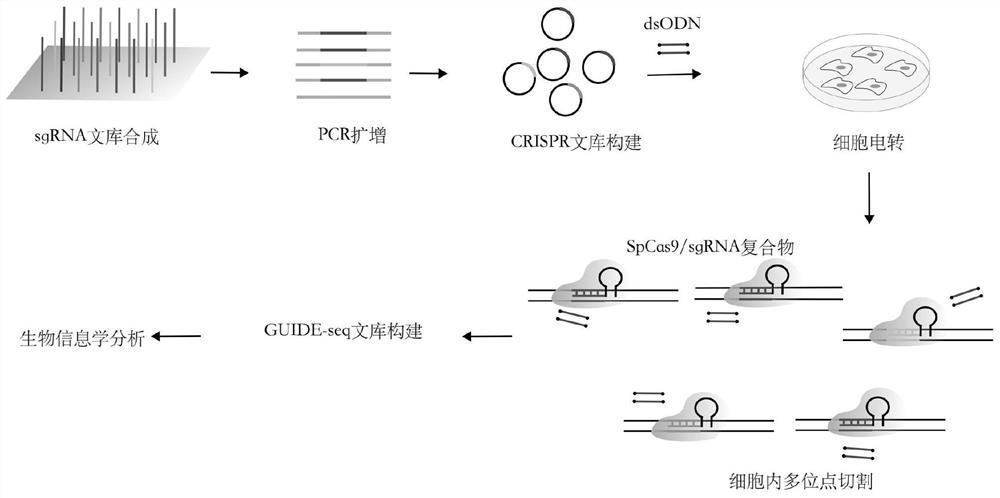
[0019] Specific Embodiment 1: In this embodiment, an accurate method for evaluating in vivo target activity and off-target detection in batches can be carried out according to the following steps:
[0020] 1. Design and synthesize sgRNA library oligonucleotide fragments for target genes;
[0021] 2. PCR amplifies the sgRNA library, and then purifies it to obtain the amplified and purified sgRNA library;
[0022] 3. Cloning the amplified and purified sgRNA library onto the SpCas9 protein expression plasmid to construct a CRISPR library;
[0023] 4. Electrotransfer of CRISPR library with dsODN tag into cells;
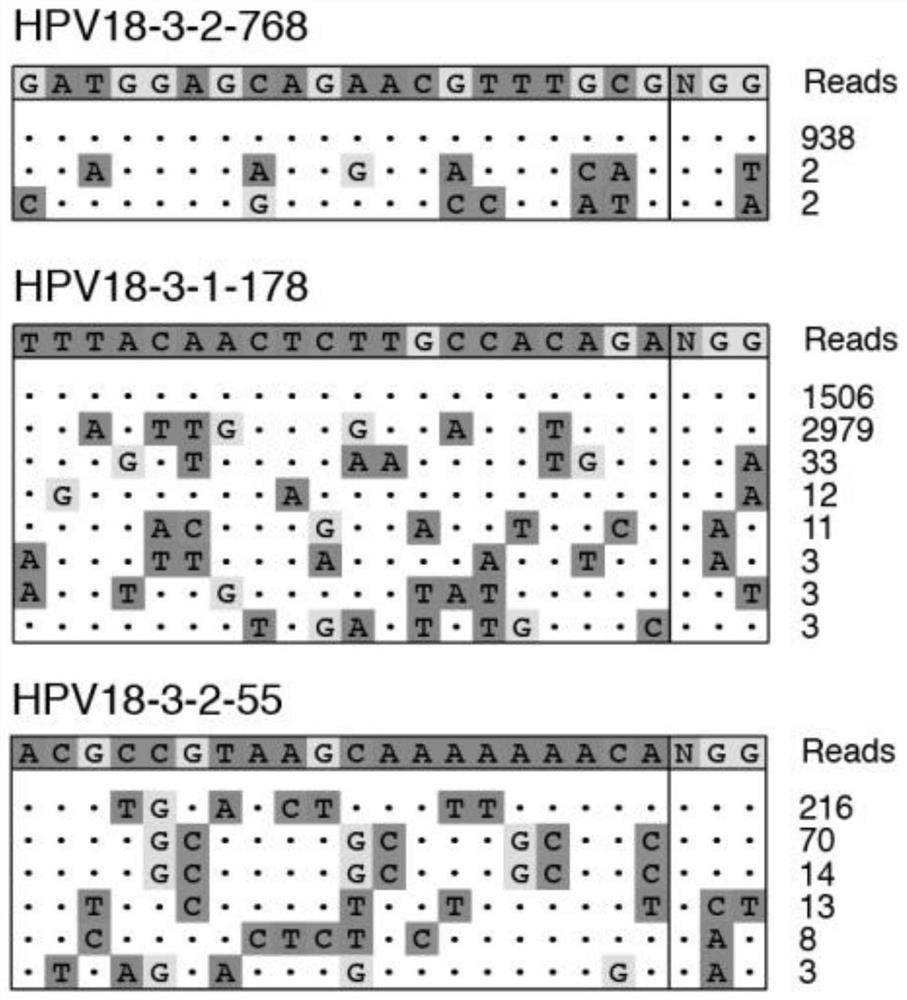
[0024] 5. Three days after transfection, cells were collected to extract DNA, and GUIDE-seq library construction was performed;
[0025] Sixth, conduct bioinformatics analysis to obtain the specific on-target activity and off-target situation of each sgRNA, that is, to complete the method of on-target activity evaluation and off-target detection.
[0026] The beneficia...
specific Embodiment approach 2
[0030] Embodiment 2: This embodiment differs from Embodiment 1 in that: in step 1, CRISPRRGEN software is used to design sgRNA targeting the full length of the target genome, and multiple sgRNAs are synthesized by primers or chip synthesis. Others are the same as the first embodiment.
specific Embodiment approach 3
[0031] Specific embodiment three: the difference between this embodiment and specific embodiment one or two is: the reaction system and reaction program of PCR in the step 2 are as follows:
[0032]
[0033]Reaction program: 98°C for 45s; 20 cycles of (98°C for 15s, 60°C for 30s, 72°C for 2min), 72°C for 2min, 4°C; PCR product was purified after PCR was completed. Others are the same as one of the specific embodiments 1 to 3.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com