HER2 gene fluorescent in-situ hybridization probe as well as preparation method and application thereof
A fluorescence in situ hybridization and probe technology, applied in biochemical equipment and methods, DNA preparation, DNA/RNA fragments, etc., can solve the problem of uncertain number of fluorescent probes, low probe specificity, and lack of small fragmentation etc. to achieve the effects of reducing background interference, increasing hybridization efficiency, and reducing hybridization time
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
preparation example Construction
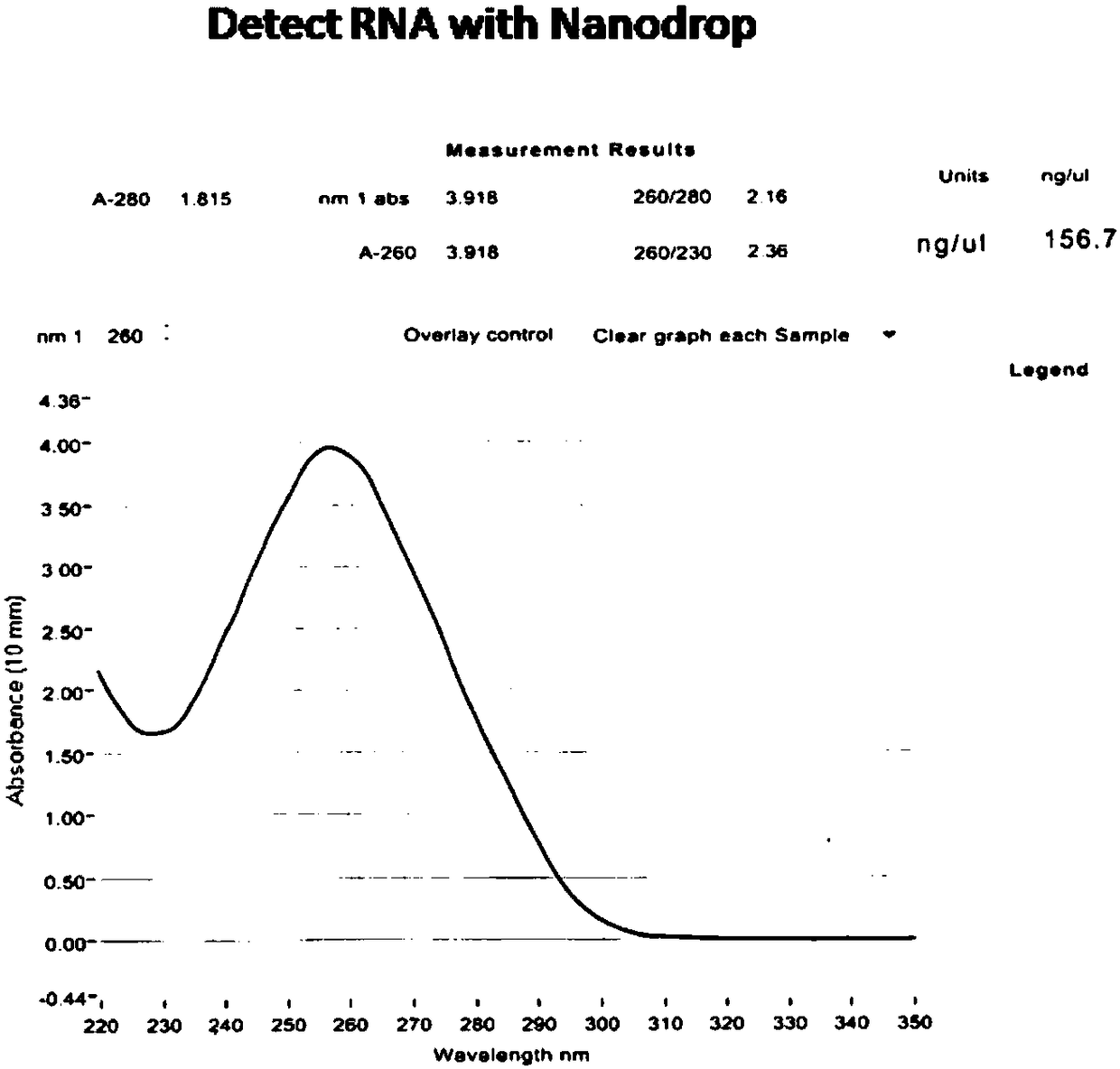
[0029] The preparation method of the HER2 gene fluorescence in situ hybridization probe of one embodiment, comprises the steps:
[0030] Step 1: Construction of the probe library: Select the non-repetitive target gene region on chromosome 17 of the human genome containing the HER2 gene, and construct a single-stranded fragment of 35nt-200nt in length that is completely complementary to the target gene region every 1-10bp as Candidate probes are compared by BLAST to obtain the Tm value of non-specific hybridization, remove the non-specific Tm value for candidate probes at 45°C, and select non-overlapping candidate probes. Add the amplification primer fragments to obtain the target probe library.
[0031] The target gene region in this embodiment is a non-repeating region of the genome, and by designing candidate probes for the non-repeating region, non-specific reactions and background interference can be reduced.
[0032] For different target gene regions, construct candidate...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com