Structure-based nucleic acid aptamer optimization design method
A nucleic acid aptamer and optimized design technology, which is used in the analysis of two-dimensional or three-dimensional molecular structures, genomics, instruments, etc., can solve problems such as difficult mass application, and achieve the effect of improving affinity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
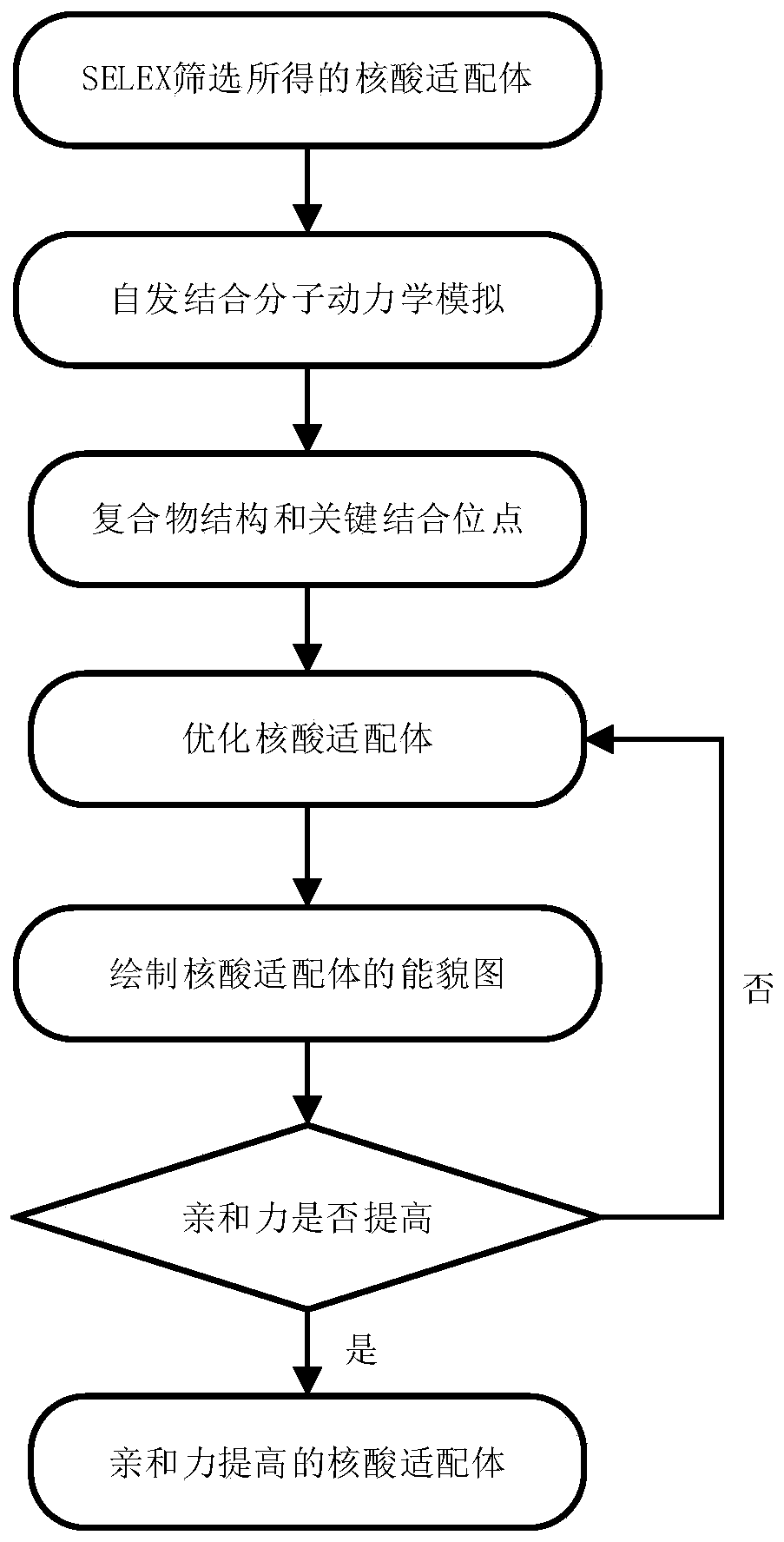
[0027] The specific process of the method of the present invention will be further described below by taking the optimal design method of the G-quadruplex nucleic acid aptamer GO18-T-d targeting GTX1 / 4 (gonyautoxin 1 / 4) as an example.
[0028] Step 1: Perform a spontaneous binding molecular dynamics simulation
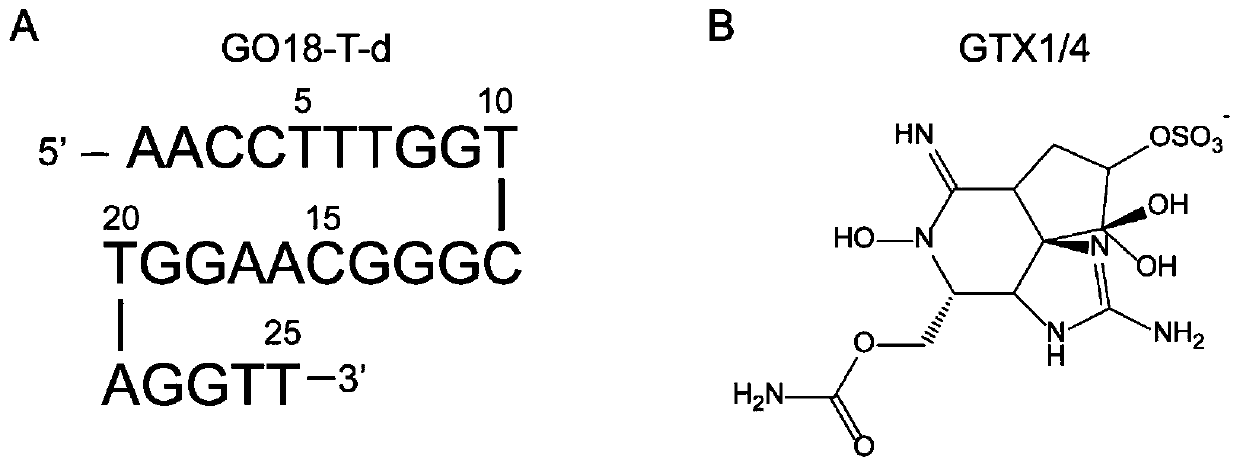
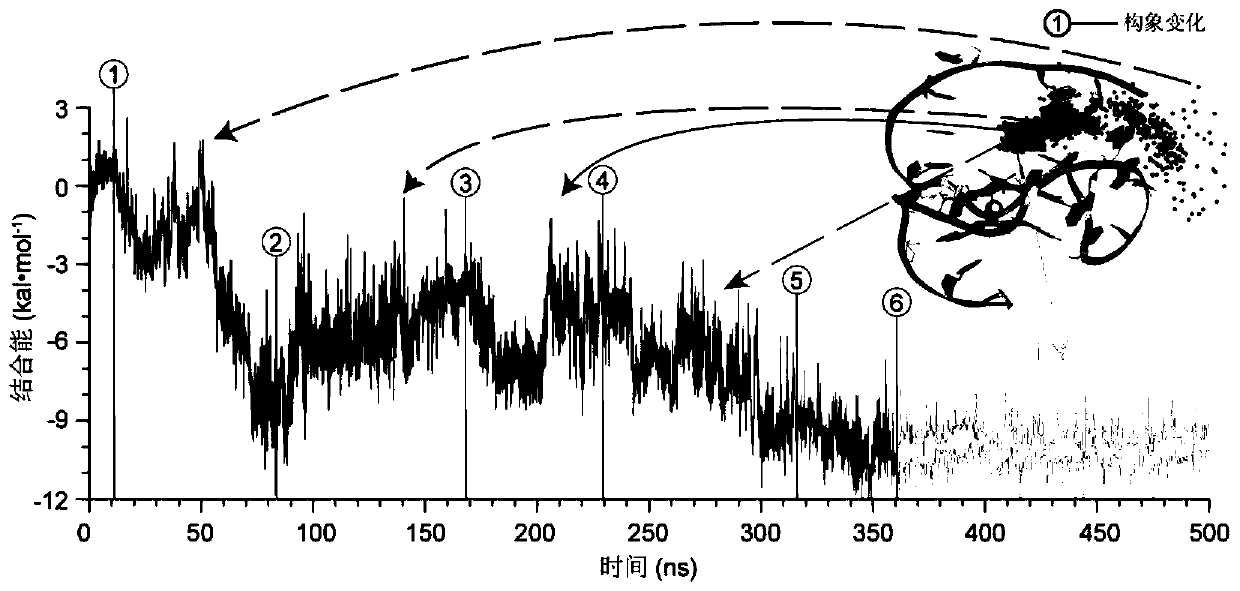
[0029] The first step is an unbiased spontaneous binding simulation with the goal of obtaining a stable and plausible 3D structure of the GO18-T-d:GTX1 / 4 complex. The sequence of the nucleic acid aptamer GO18-T-d and the structure of the toxin small molecule GTX1 / 4 are as follows figure 2shown. The structure of GO18-T-d was generated by the AutoPSF plugin in 3D-Nus(13) and VMD(14), and screened by temperature-dependent molecular dynamics simulations. The GAFF parameters of the toxin molecule GTX1 / 4 were generated using the Antechamber software package that comes with AmberTools. After obtaining the force field parameters of the small molecule, in order to avoid the...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com