Method for detecting full-length sequence of circular RNA
A detection method and DNA sequence technology are applied in the field of circular RNA detection to achieve the effect of quick and easy detection
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0044] 1. Circular RNA primer design principle:
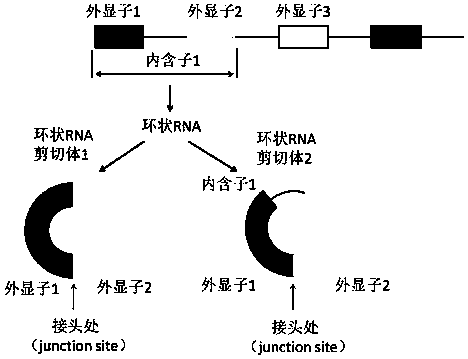
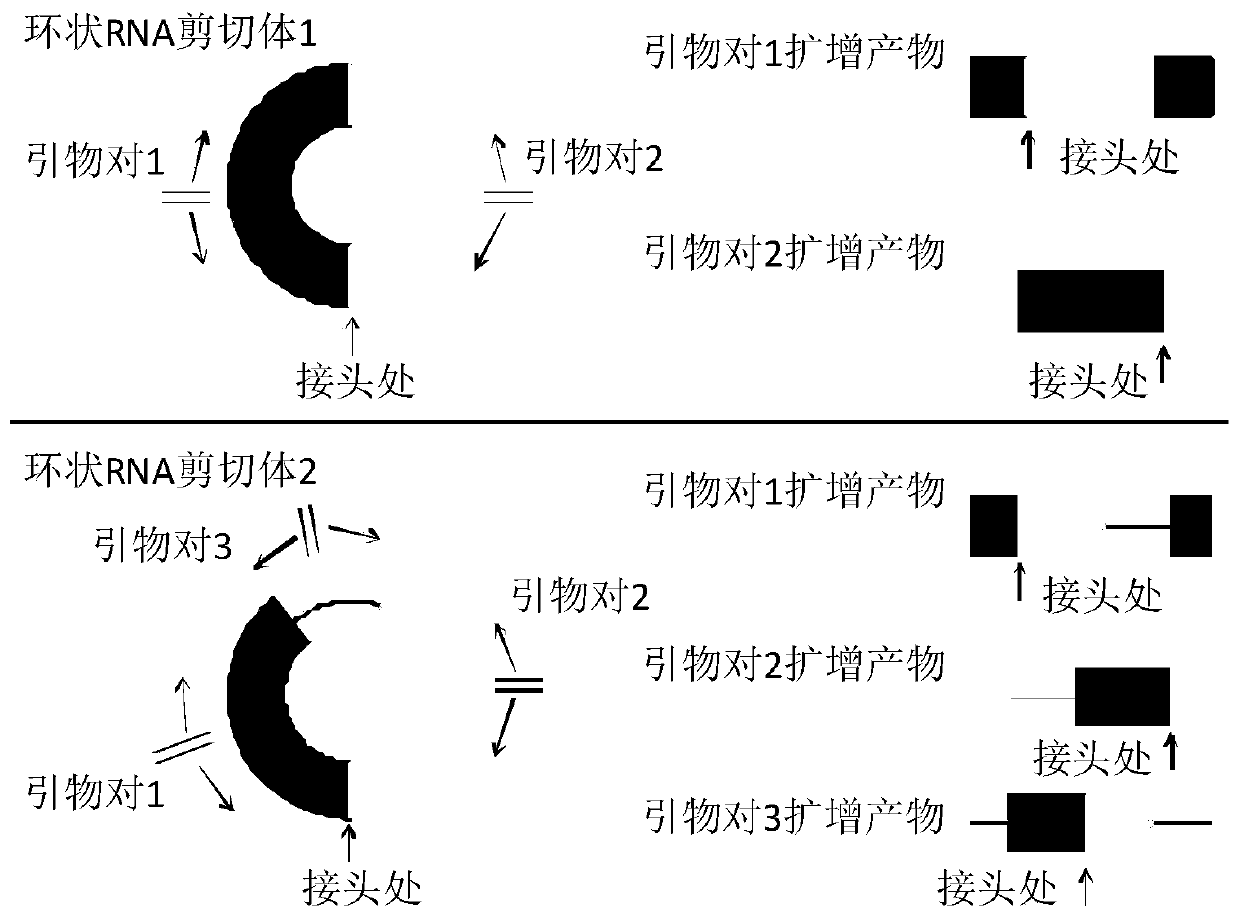
[0045] like figure 1As shown, if the primers directed at several bases of the sequence at the joint of the circular RNA are conventionally designed, only the sequence at the joint can be amplified due to PCR amplification. But for circular RNA, the circular RNA can form alternative cuts during the formation process, subtract or retain the intron, such primers can only detect the sequence at the junction, but cannot detect the circular RNA full-length sequence. In summary, according to the characteristics of circular RNAs, we simply and quickly designed primers for different alternative splicing modes of circular RNAs and capable of detecting the full-length circular sequences.
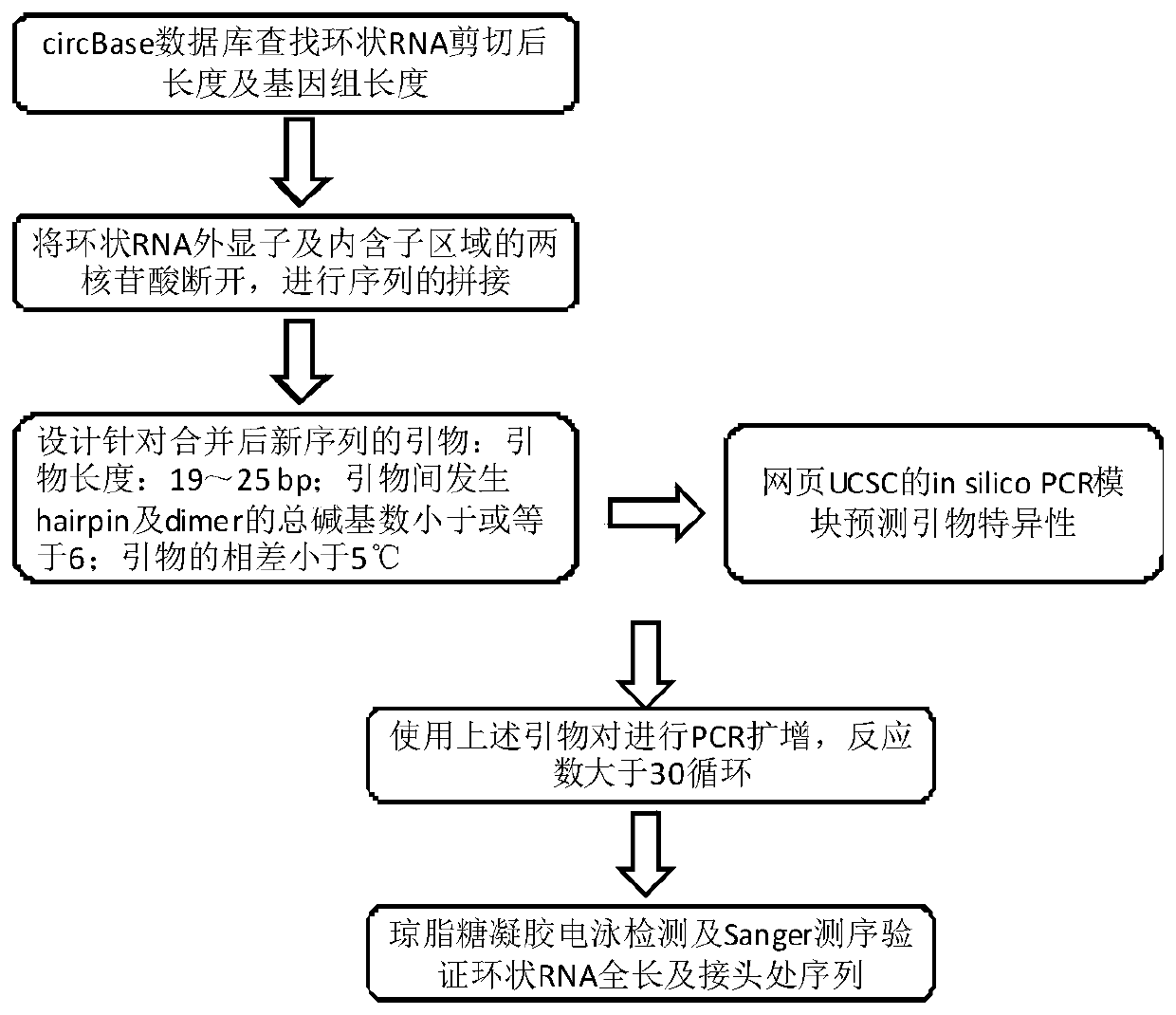
[0046] 2. Circular RNA primer design scheme of the present invention:
[0047] (1) Take hsa_circ_0007766, hsa_circ_0001355 and hsa_circ_0001538 as examples respectively. Find the genomic DNA sequence of the circular RNA in circBase (where English up...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com