Drug relocation model based on pathogenic contribution network analysis
A network analysis and relocation technology, applied in the fields of computer technology and biomedicine, to alleviate the difficulty of research and development and reduce the scope
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
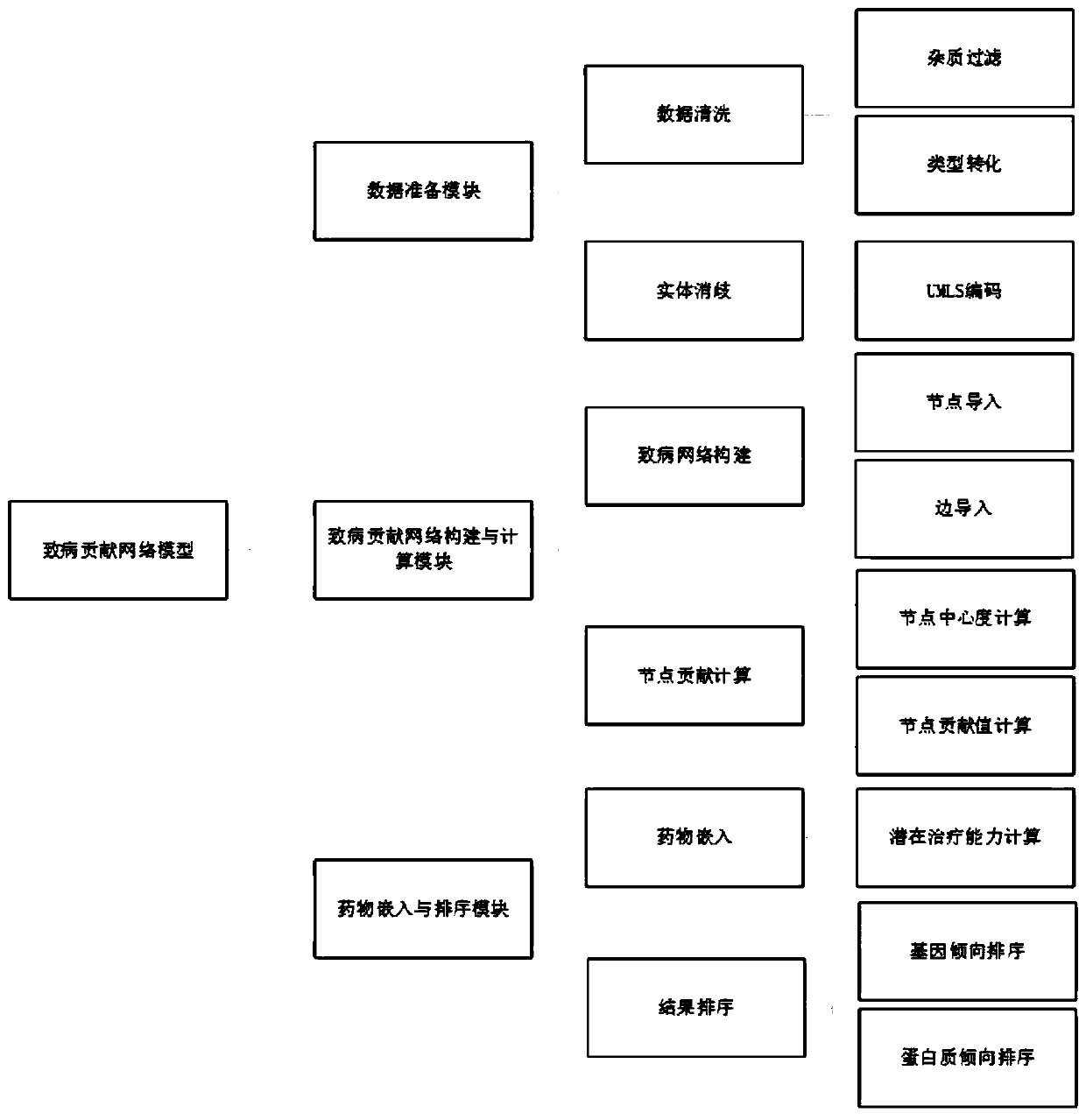
[0039] The present invention is a drug relocation model based on network analysis of pathogenic contribution.
[0040] Implementation technology: In the process of model construction and system development, the java programming language is used as the background language, the database uses the Mysql relational database and the Neo4j graph database, and the query uses SQL+CQL (Cypher). The display part was processed and rendered using the neo4j Bloom tool. The calculation part uses the neo4j APOC stored procedure and graph algorithm package.
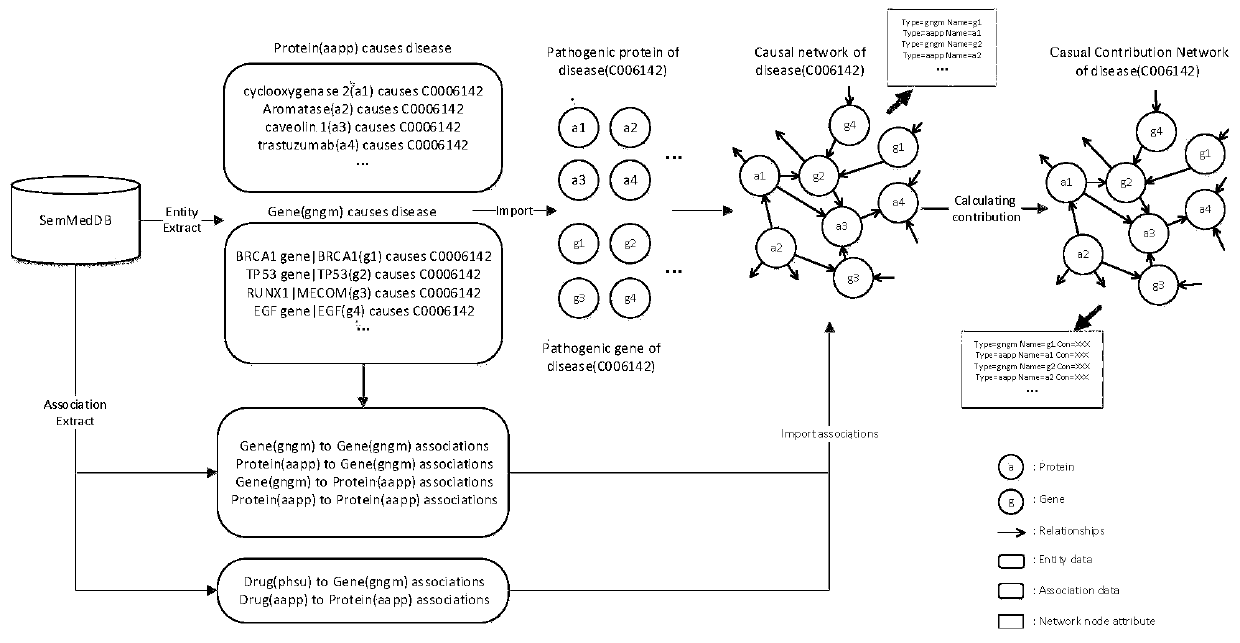
[0041] The model can conduct drug repositioning research on any disease, select Parkinson's disease (PD) as the target disease, and select the SemmedDB database as the data source for case presentation.
[0042] Data preparation module:
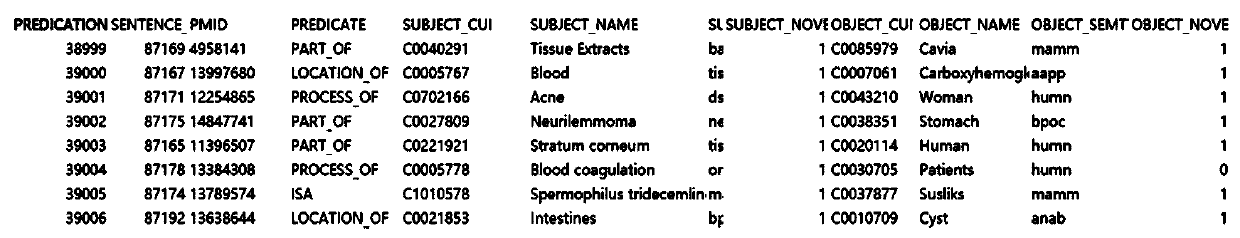
[0043] Selecting SemmedDB as the data source, extracting all PD-related genes and proteins from SemmedDB and the relationship between them are unified into the triple data format of subject-predicate-o...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com