Primer for identifying rhizoma atractylodis medicinal species by using chloroplast genes, and applications thereof
A technology of chloroplast and Atractylodes genus, which is applied in the field of using chloroplast genes to identify primers for medicinal species of Atractylodes genus, which can solve the problems of unsatisfactory performance, unclear species boundaries, and low resolution
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
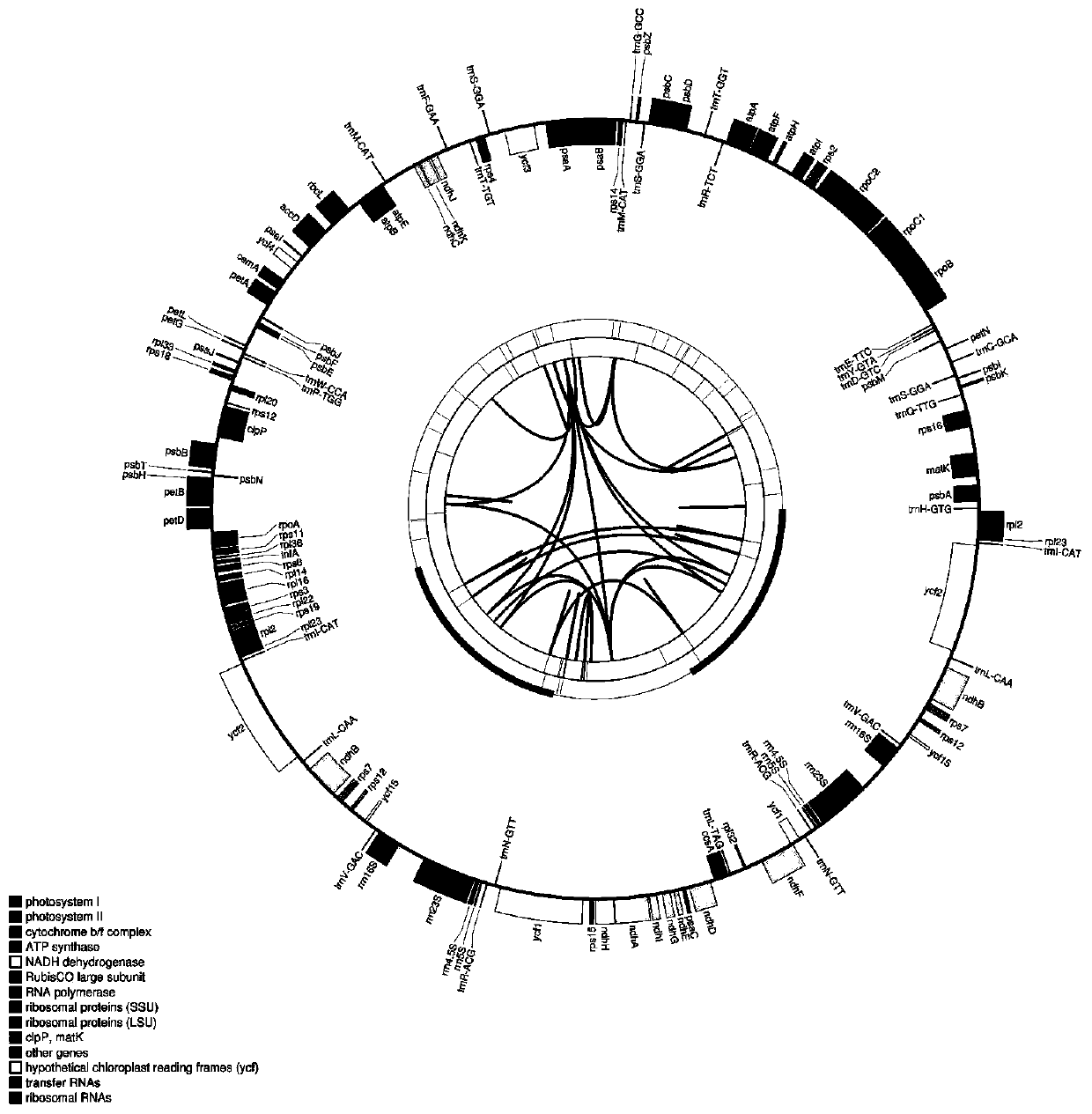
[0044] 1. The genomes of Atractylodes Rhizoma Atractylodes Rhizome, Atractylodes Rhizoma Atractylodes Rhizome and Atractylodes Rhizoma Atractylodes were extracted, sequenced by NGS technology, and the chloroplast genome sequences of Atractylodes Rhizome, Atractylodes Rhizome and Atractylodes Rhizome were obtained, and data analysis was performed on the sequences by software.
[0045] Data difference analysis method:
[0046] The present invention uses REPuter software to identify four types of repeat sequences, including forward repeats, palindromic repeats, inverted repeats and complementary repeats; the minimum repeat unit size is set to 30bp, and the critical value of similarity between repeat units is set to 90%.
[0047] Use MISA software to predict simple repeats (SSR), and the thresholds are as follows: ① When the number of repeating unit nucleotide residues is 1, the number of repeating units is at least 8; ② The number of repeating unit nucleotide residues is 2-3 3, ...
Embodiment 2
[0093] In order to further verify the reliability of the above molecular markers, Atractylodes Rhizoma Atractylodes Rhizome and Atractylodes Rhizoma Atractylodis selected plant materials from 3 origins, and 6 individuals (Table 10) were collected from each origin. Atractylodes macrophylla selected plant materials from 2 origins, and collected 2 individuals (Table 10) from each origin. Total DNA extraction was performed on all 40 individuals. PCR amplification was performed with the primer pair cz11_F / cz11_R. The corresponding PCR products were sequenced by Sanger sequencing. The alignment results of the 40 PCR product sequences are shown in ( Figure 5 )middle. The 36 individuals of Atractylodes Rhizoma Atractylodes Rhizome and Atractylodes Rhizoma Atractylodes Rhizome all showed expected marker sequences (Table 10) and were successfully verified. The 4 individuals of Atractylodes macrophylla also showed the expected marker sequence (Table 10) and were successfully verifie...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com