Pyrophosphorolysis activated fluorescence to measure PAP amplification of nucleic acid
A pyrophosphorylation and activation technology, applied in the field of molecular biology, can solve the problem of inability to identify multiple template amplification and other problems
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
example 1
[0138] Example 1: Materials and methods
[0139] Preparation of primers
[0140] The 3'ddCMP blocking primer used was chemically synthesized in the 3'-5' direction by Integrated DNA Technologies and purified by HPLC.
[0141] The 3'ddAMP, ddTMP and ddGMP blocking primers are synthesized by adding ddATP, ddTTP and ddGTP to the 3'end of the deoxy-oligonucleotide by terminal transferase (Liu and Sommer, 2000; Liu and Sommer, 2002).
[0142] The primers with single labeling (FAM or HEX) at the 5'end or internal region are chemically synthesized along the 3'-5' direction by Integrated DNA Technologies, and purified by HPLC.
[0143] Rhodamine dye-labeled dideoxynucleotide analogs or blockers, TAMRA-ddATP, TAMRA-ddUTP, TAMRA-ddGTP and TAMRA-ddCTP, purchased from PerkinElmer Life Sciences, in which the quenching group TAMRA is covalently linked to The base of the dideoxydeoxynucleotide of the primer. Then they are added to the 3'end of the single-labeled primers by terminal transferase to sy...
example 2
[0154] Example 2: Single and multiple PAP testing of GNAS and HIV genes
[0155] Single-plex and multiple PAP tests of GNAS and HIV genes are used to demonstrate how pyrophosphorylation-activated fluorescence works.
[0156] The GNAS and HIV genes were selected as targets and controls. Because HIV-1 virus DNA can be integrated into the human genome, the use of blood leukocyte genomic DNA for testing has become a medical requirement.
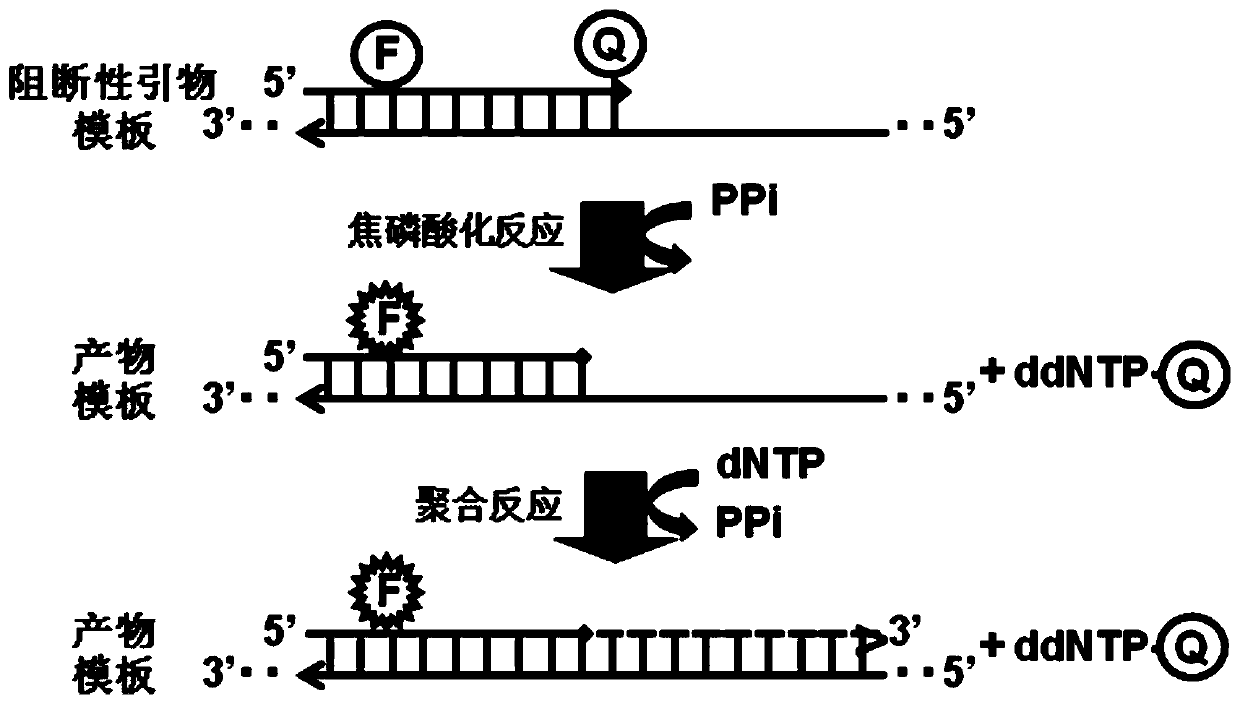
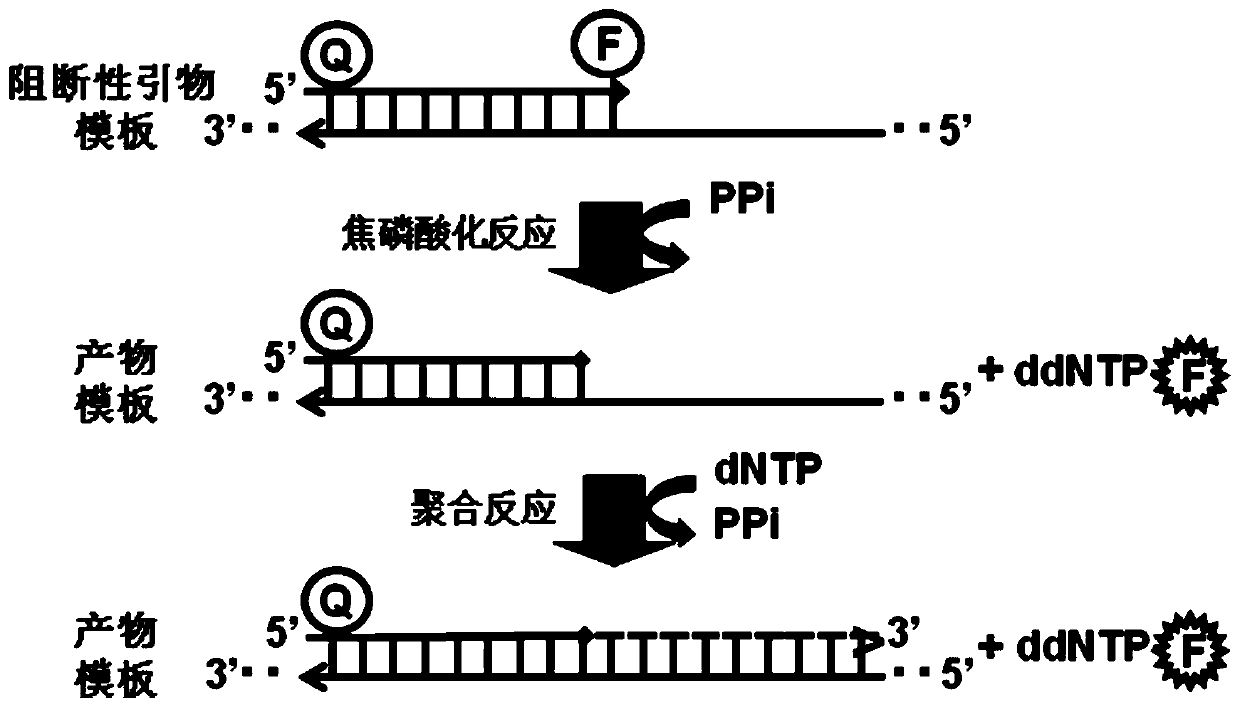
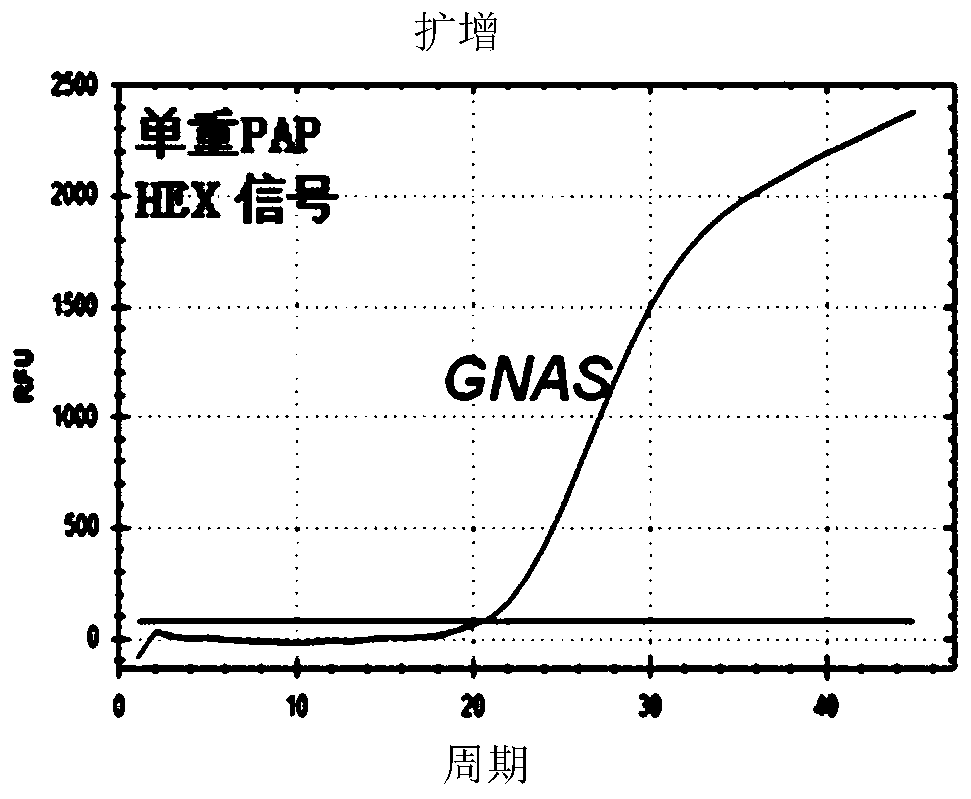
[0157] A single-plex PAP detects the GNAS gene using a forward fluorophore-quencher double-labeled blocking primer (SEQ ID 1) and a reverse blocking primer (SEQ ID 2) (Table 2). Fluorophore-quenching group double-labeled blocking primer (SEQ ID 1) with a HEX fluorophore covalently linked to the 5'end dAMP of the primer and a covalent bond to the 3'end ddCMP The TAMRA quencher on the group. When the GNAS gene was amplified from 100,000 copies of human wild-type genomic DNA (i.e., 330ng), a HEX fluorescence signal was generated, and the number of Ct 20...
example 3
[0167] Example 3: Single and multiple delayed PAP detection of GNAS and HIV genes
[0168] Delayed PAP was invented by introducing artificial mutations into the 3'region of the blocking primer, which can delay product accumulation to a later time or cycle during PAP amplification (US Patent Application No. 62687325). Pyrophosphorylation activated fluorescence was further confirmed in this delayed PAP test.
[0169] A single delayed PAP detects the GNAS gene using a forward fluorophore-quenching group double-labeled blocking primer (SEQ ID 1) (Table 2) and a 5th nucleotide from the 3'end There is a reverse M1 blocking primer with artificial G to T mutation (SEQ ID 6) (Table 3). Another single delayed PAP detects the GNAS gene using a forward fluorophore-quenching group double-labeled blocking primer (SEQ ID 1) and an introduction at the 3rd nucleotide from the 3'end A reverse M2 blocking primer with C to A worker mutation (SEQ ID 7) (Table 3). When using 100,000 copies of human w...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com